Long-chain non-coding RNA (Ribonucleic Acid) correlated to occurrence and development of human hepatocellular carcinoma, amplification detection method and application
A long-chain non-coding, human hepatocellular carcinoma technology, applied in biochemical equipment and methods, microbial assay/inspection, DNA/RNA fragments, etc., can solve problems such as biological functions and molecular mechanisms that remain to be elucidated
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
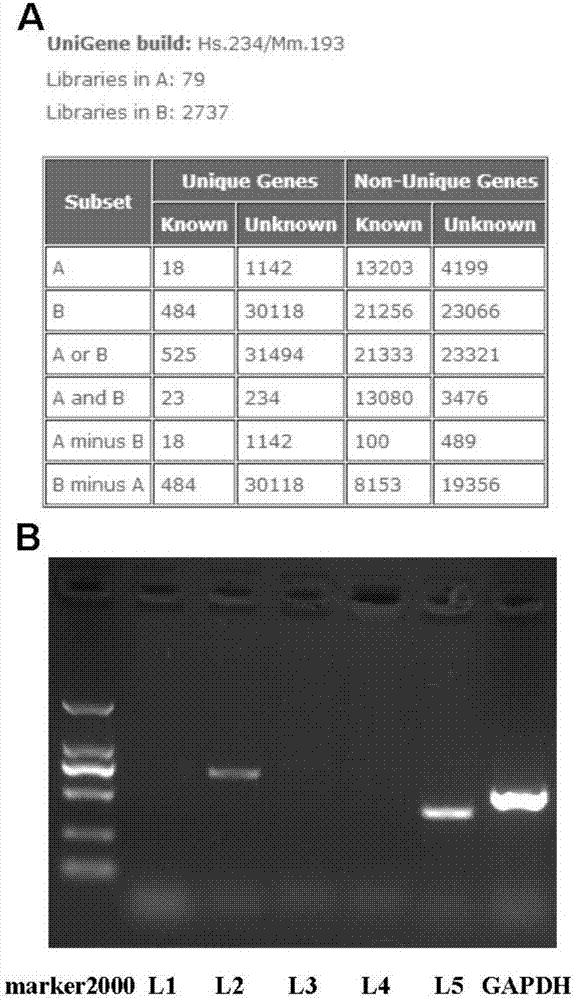
[0033] Example 1: Preliminary screening of new genes related to hepatocellular carcinoma by bioinformatics
[0034] The cDNA xProfiler of the Cancer Genome Analysis Project CGAP platform was used together with the BLAST and DDD (Digital Differential Display) tools of the National Center for Biotechnology Information NCBI platform.
[0035] (1) Use the cDNA xProfiler tool of the CGAP platform:
[0036] Pool A selects all cDNA libraries related to liver cancer;
[0037] Pool B selects all cancer and non-cancer cDNA libraries except liver cancer.
[0038] (2) Select 1142 unknown EST sequences in the differential comparison results of the cDNA library, compare them in the Genbank database, exclude known repetitive genes and genome contamination sequences, and obtain 5 EST sequences that can be used for the next step of screening, Excluding the fragments with no specific bands amplified by PCR in the 3 EST sequences, leaving HCCL2 and HCCL5 for comparison, it was found that HCCL5...
Embodiment 2
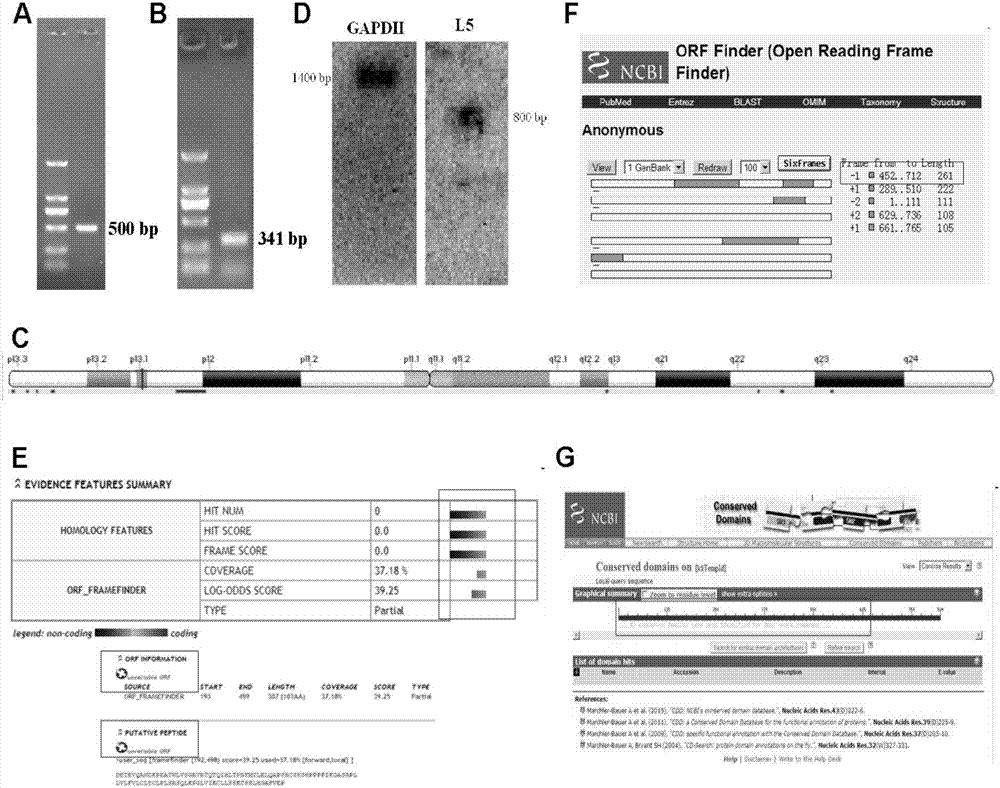
[0039] Embodiment 2: PCR amplification full-length gene
[0040] 1) Preparation and extraction of human hepatocellular carcinoma HepG2 cell RNA;
[0041] 2) Using RNA as a template to synthesize a 3'RACE cDNA template, use the following primers to perform PCR to obtain a 3'RACE product;
[0042] The 3'RACE primer 3L3 nucleotide sequence of HCCL5 is as SEQ ID NO.2 in the sequence listing;
[0043] 3'RACE nested PCR primer of HCCL5: 3L3N nucleotide sequence as SEQ ID NO.3 in the sequence listing
[0044] 3) Synthesize 5'RACE cDNA template using RNA as a template, and use the following primers to perform PCR to obtain 5'RACE products;
[0045] The 5' RACE primer 5L3 nucleotide sequence of HCCL5 is as SEQ ID NO.4 in the sequence listing;
[0046] The 5' RACE nested PCR primer 5L3N nucleotide sequence of HCCL5 is as SEQ ID NO.5 in the sequence listing;
[0047] 4) By splicing 5'RACE and 3'RACE sequences, the full-length sequence was obtained.
[0048] According to the conventi...
Embodiment 3
[0049] Example 3: Obtaining the full length of lncRNA HCCL5 gene and construction of eukaryotic expression vector
[0050] Design PCR primers to amplify the full length of lncRNA HCCL5, add Hind III and Xho I restriction sites to the primers respectively, and connect the obtained fragments to the pcDNA3.1(+) eukaryotic expression vector, see nucleosides for the nucleotide sequence of the amplification primers Acid sequence list SEQID NO.2, NO.5. The amplified fragments were inserted into the pcDNA3.1(+) eukaryotic expression vector after conventional digestion, ligation and other operations. The obtained recombinant plasmid was confirmed by DNA sequencing, and the result showed that the recombinant plasmid was correct and could be used for follow-up research.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com