miRNA data and tissue specificity network-based drug reorientation method
A tissue-specific and repositioning technology, applied in the field of bioinformatics, can solve the problem of low accuracy of prediction results and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0033] The present invention will be further described in detail below in conjunction with the accompanying drawings and specific embodiments.
[0034] This embodiment takes breast cancer as an example.
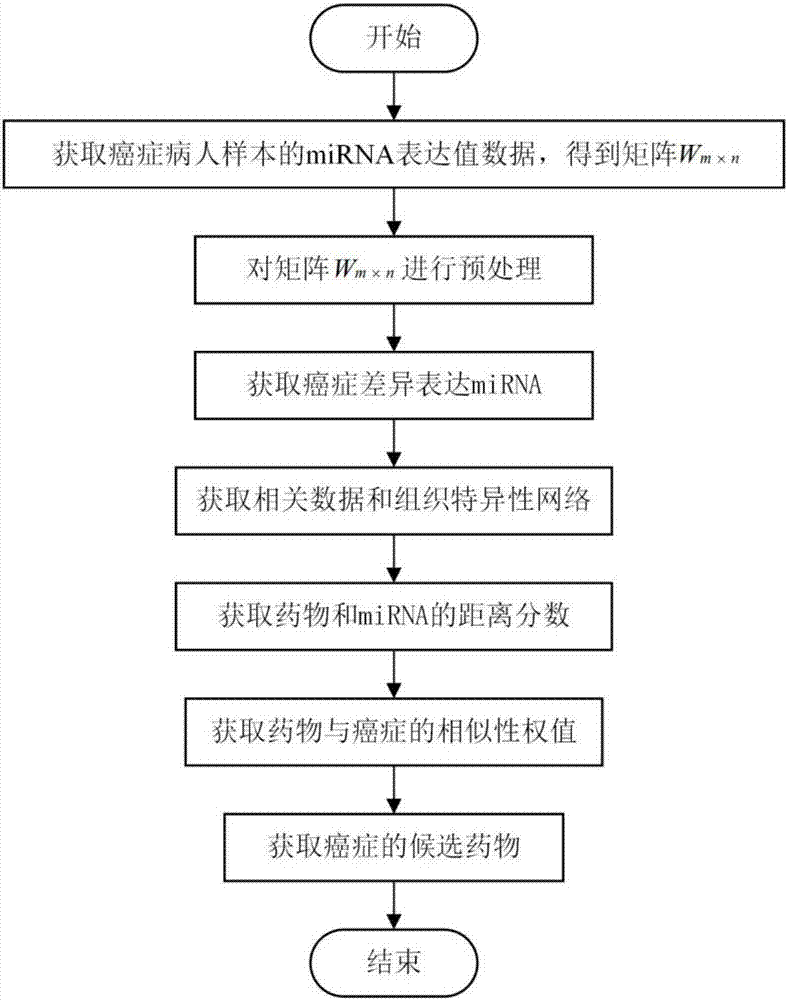
[0035] refer to figure 1A drug repositioning method based on miRNA data and a tissue-specific network, comprising the steps of:
[0036] Step 1) download the miRNA expression value data of 1189 breast cancer patient samples from the TCGA database, and obtain matrix W, and the W matrix has 503 rows and 1189 columns, wherein the rows represent 503 miRNAs, and the columns represent 1189 breast cancer patient samples;
[0037] Step 2) Preprocess the matrix W: calculate the mean value of each row of miRNA in the matrix W, and use it as the expression value R of the row of miRNA i , get a vector r=[R that contains 503 miRNA expression values 1 , R 2 , R 3 ,...,R i ,...,R 503 ], wherein, i represents the index of miRNA, and i=1,2,3,...,503;
[0038] miRNA expression value R ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com