Differential methylation site identification method
An identification method and methylation technology, applied in the fields of genomics, special data processing applications, instruments, etc., to achieve the effect of stable model, better performance, and better performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0036] For the convenience of description, the relevant technical terms appearing in the specific implementation are explained first:
[0037] fastDMA (Fast Differential MethylationAnalysis): Rapid differential methylation analysis method
[0038] ChAMP (The Chip Analysis Methylation Pipeline): Methylation chip analysis
[0039] TCGA (The Cancer Genome Atlas): Cancer Genome Atlas;
[0040] SWAN (Subset-quantile Within Array Normalization): Subset quantile normalization method;
[0041] ComBat(Empirical Bayes methods): Empirical Bayesian method;
[0042] ESCA (esophageal carcinoma): Esophageal cancer.
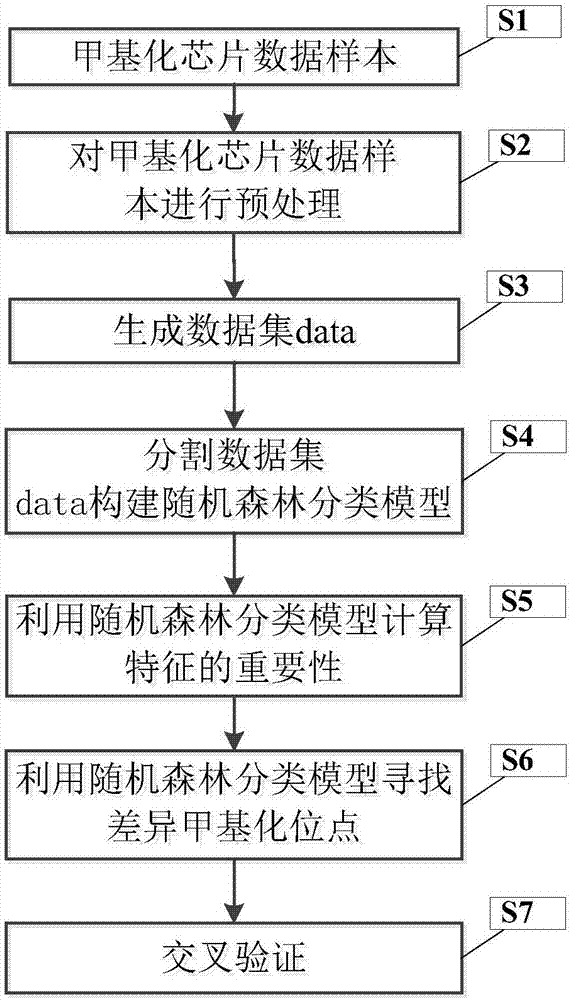
[0043] figure 1 It is a flowchart of a differential methylation site identification method of the present invention.
[0044] In this example, if figure 1 As shown, a differential methylation site recognition method of the present invention comprises the following steps:
[0045] S1. Randomly obtain N groups of 450K methylation microarray data samples of a cancer from the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com