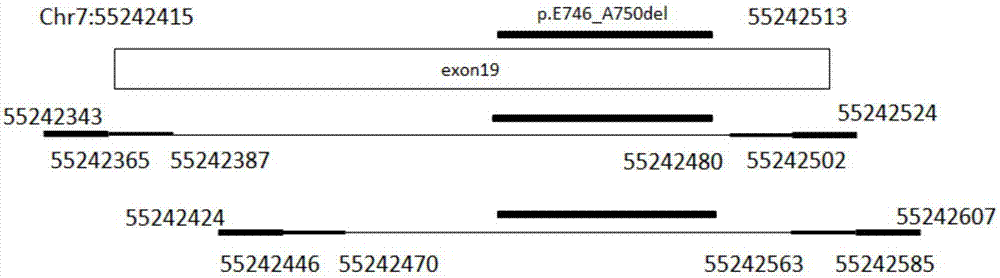
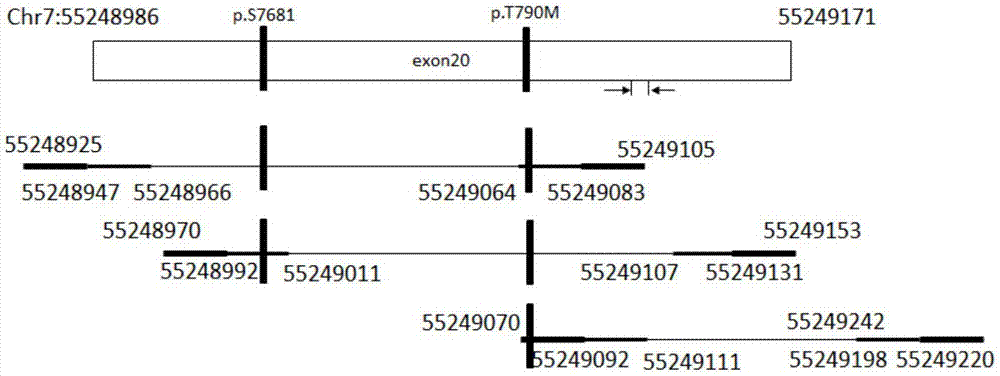
Primer composition, kit and method for detecting EGFR gene mutations
A primer composition and composition technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/testing, etc., can solve problems such as high cost and cumbersome operation, and achieve the effect of improving convenience and flexibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] The specific library construction method of the application is as follows:
[0060] (1) The first round of PCR
[0061](1) The first round of multiplex PCR amplification: A single portion needs to be amplified simultaneously according to the PCR amplification reaction systems of library A and library B (as shown in Table 4 and Table 5 below, respectively), (25 μl system):
[0062] Table 4:
[0063] A library component (1╳)
Volume (μl)
PCR Enzyme Mix
16.5
Primer Mix A
2
template DNA
X
Nuclease-free ultrapure water
6.5-X
[0064] Attachment: Primer mixture A is a mixture of SEQ ID NO:1-10.
[0065] table 5:
[0066] B library components (1╳)
Volume (μl)
PCR Enzyme Mix
14.5
Primer Mix B
2
template DNA
X
Nuclease-free ultrapure water
8.5-X
[0067] Attachment: Primer mixture B is a mixture of SEQ ID NO: 11-18.
[0068] (2) PCR amplification, put each reac...
Embodiment 2
[0090] Purchase traceable positive cell lines or FFPE samples of 6 EGFR gene mutation sites and EGFR gene mutation-negative cell lines from the market. Or FFPE DNA and EGFR gene mutation-negative cell line DNA are mixed in a certain proportion to make positive reference substances with different mutation frequencies. The specific information of the positive reference substances used in the following examples of this application is shown in Table 8:
[0091] Table 8:
[0092]
[0093] The method of Example 1 was used to detect the reference products of different positive mutation types, and the detection was repeated 6 times (L1-L6). The test results are shown in Table 9 below. The results showed that only the corresponding mutation types were positive, and the coincidence rate was 100%.
[0094] Table 9:
[0095]
[0096]
Embodiment 3
[0098] The method of Example 1 was used to detect 10 reference samples (N1-N10) of negative mutation types, and the test results are shown in Table 10 below. The results showed that they were all negative (neg), and the coincidence rate was 100%.
[0099] Table 10:
[0100]
[0101]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com