3,7-Dihydroxytropolone Biosynthetic Gene Cluster and Its Application
A technology for hydroxytropolone and biosynthesis, which is applied in the field of microbial gene resources and genetic engineering, can solve the problems such as the biosynthesis pathway is not well revealed, the research progress is limited, and the yield is limited.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0101] Embodiment 1, construction of Streptomyces sp.B222 cosmid library
[0102] (1) Inoculate fresh spores of Streptomyces sp.B222 into 30ml of TSBY medium (with 0.6% glycine added), and culture at 30°C and 220rpm for 16-24h.
[0103] (2) Collect the bacteria by centrifugation, weigh 0.5g of wet bacteria into a 50ml centrifuge tube, add 5ml of SET (75mM NaCl, 25mM EDTA, 20mM Tris, pH 7.5) buffer, shake to break up. Add lysozyme at a final concentration of 1mg / ml, and incubate at 37°C for 10min. After adding 1 / 10 volume of 10% SDS solution, proteinase K with a final concentration of 0.5 mg / ml was added, and incubated at 55° C. for 2 h. Add 1 / 3 volume of 5M NaCl solution, and add one volume of neutral phenolform, invert and mix at room temperature for 30min; centrifuge at 4,000rpm for 30min, draw the supernatant into another new 50ml centrifuge tube, add an equal volume of chloroform Mix again by inversion for 30 min to remove residual protein. After centrifugation at 4,0...
Embodiment 2、3
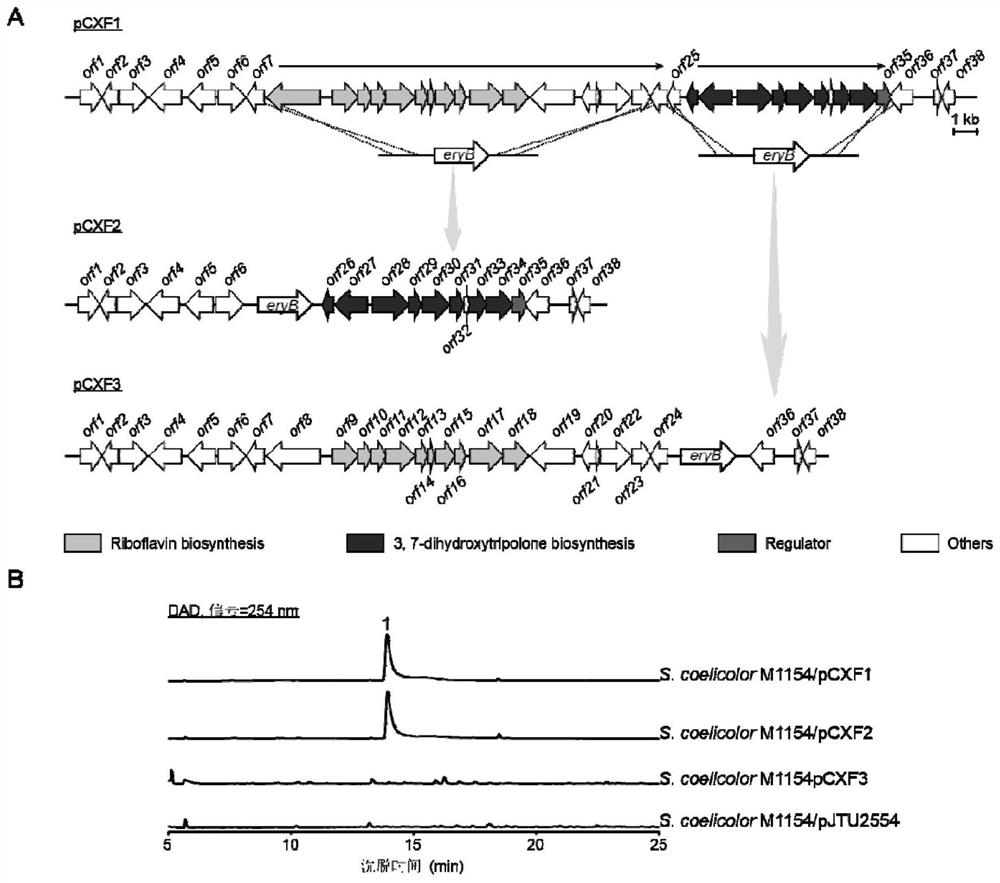
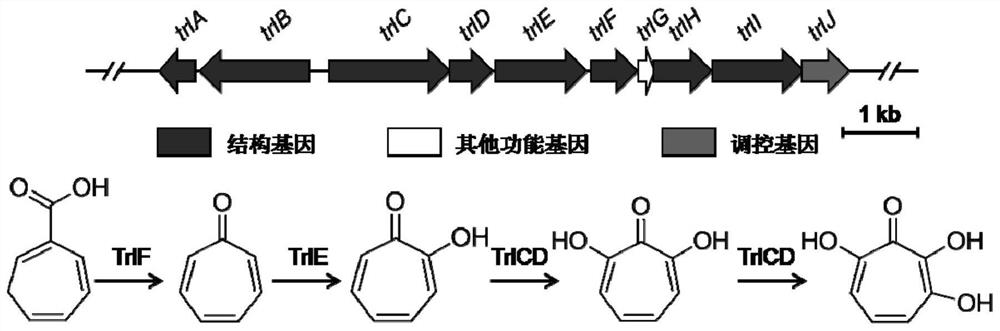
[0107] Cloning of Example 2, 3,7-Dihydroxytropolone Biosynthetic Gene Cluster
[0108] For the screening method of heterologous expression activity of Streptomyces genomic library, please refer to the literature "Chen L, Wang Y, Guo H, et al. "And literature" Xu M, Wang Y, Zhao Z, et al. Functional Genome Mining for Metabolites Encoded by Large Gene Clusters through Heterologoous Expression of a Whole-GenomeBacterial Artificial Chromosome Library in Streptomyces spp. Applied and Environmental Microbiology, 2016, 82(19) :5795-5805."
[0109] (1) Take out the cosmid library (12 pieces of 96-well plates) of Streptomyces ochraceiscleroticus B222 (ie, S. ochraceiscleroticus B222) from the -80°C refrigerator, and place it in a 37°C incubator for 2 hours until it melts completely.
[0110] (2) Add 130 μl of fresh LB medium to a 96-well plate, use a replicator to inoculate the cosmid library of Streptomyces sp.B222 into a 96-well plate, culture at 37°C, 220 rpm for 4-6 hours to OD 6...
Embodiment 3、3
[0116] Embodiment 3, 3, the heterologous expression of 7-dihydroxytropolone
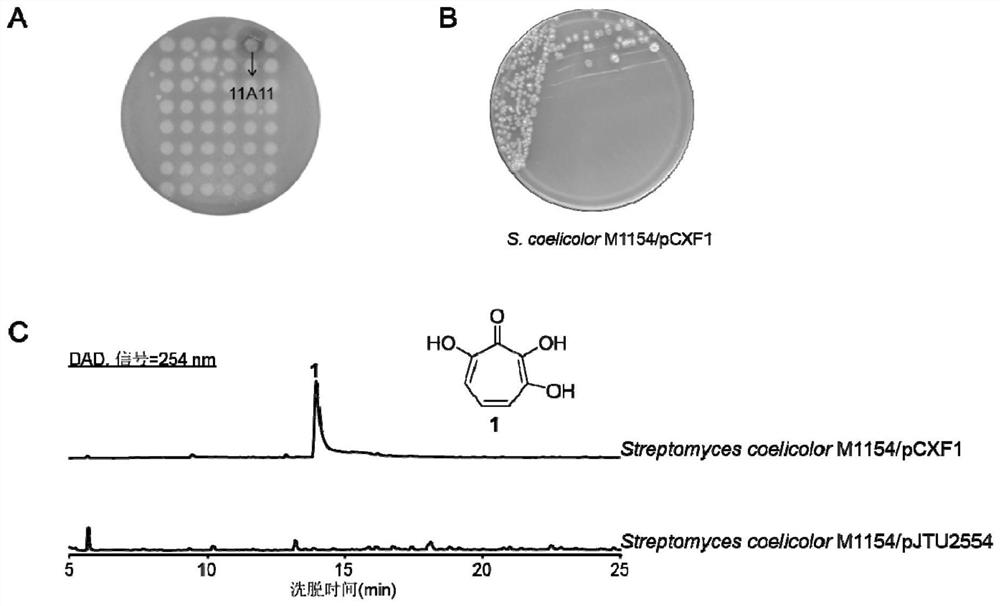
[0117] (1) Pick the 11A11 clone from the cosmid library library of Streptomyces ochraceiscleroticus B222 and put it into 3 ml of fresh LB medium (with 0.1% abramycin added), and culture overnight at 37°C and 220rpm.
[0118] (2) The cosmid DNA extracted from 11A11 was transformed into E. coli E.coli ET12567 electroporation competent cells to obtain the E.coli ET12567 / 11A11 (E.coli ET12567 / pCXF1) strain. For the convenience of description, the subsequent 11A11cosmid plasmids are all represented by pCXF1 .
[0119] (3), using the method of Escherichia coli-Streptomyces three-parent conjugation transfer, with E.coli ET12567 / pCXF1 as the donor bacterium, and S.coelicolor M1154 as the recipient bacterium, with the assistance of E.coli ET12567 / pUB307, the The pCXF plasmid was transferred into S.coelicolor M1154 to obtain the expression strain S.coelicolor M1154 / pCXF1 CGMCC No. of 3,7-dihydroxytropolone,...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap