Detection of rare gene mutations
A mutation type and mutation site technology, which is applied in the detection/testing of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0095] The technical solutions disclosed herein are further described below through examples.
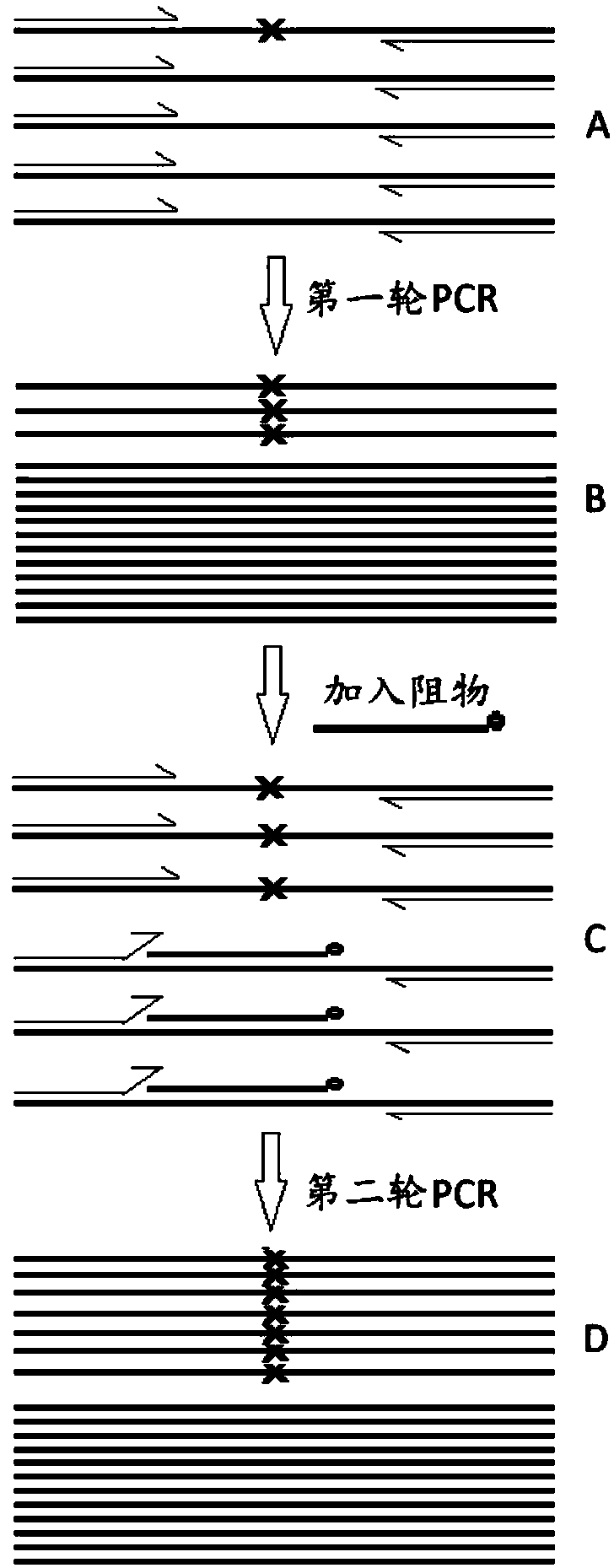
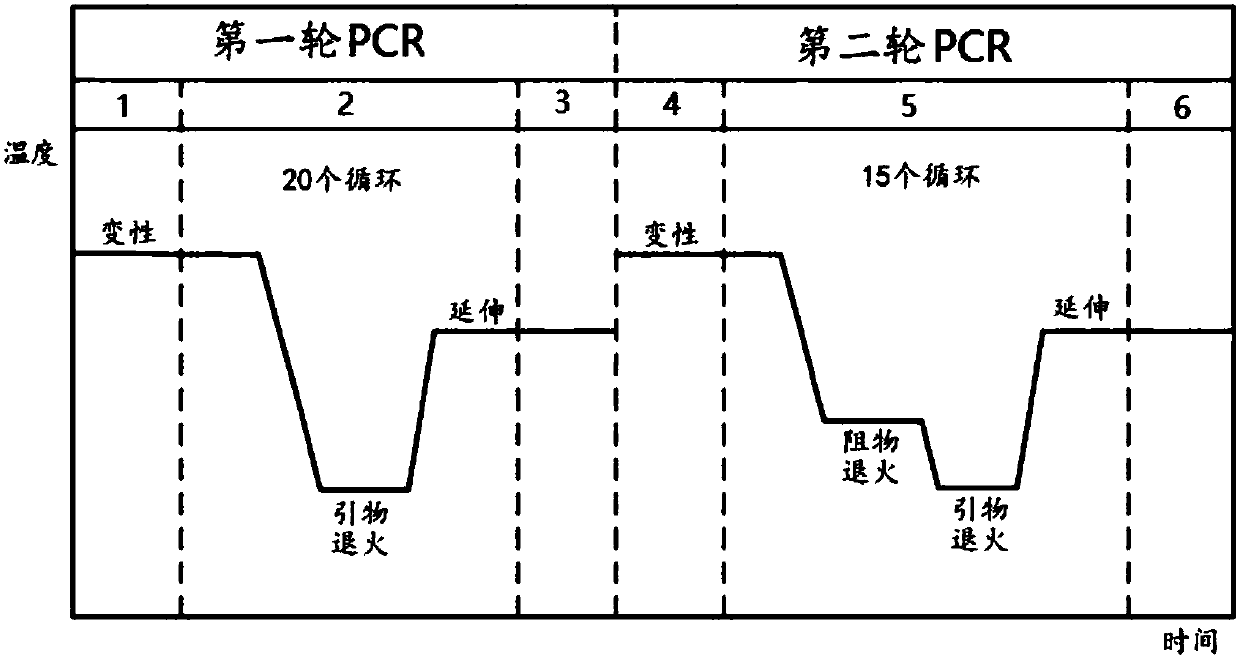
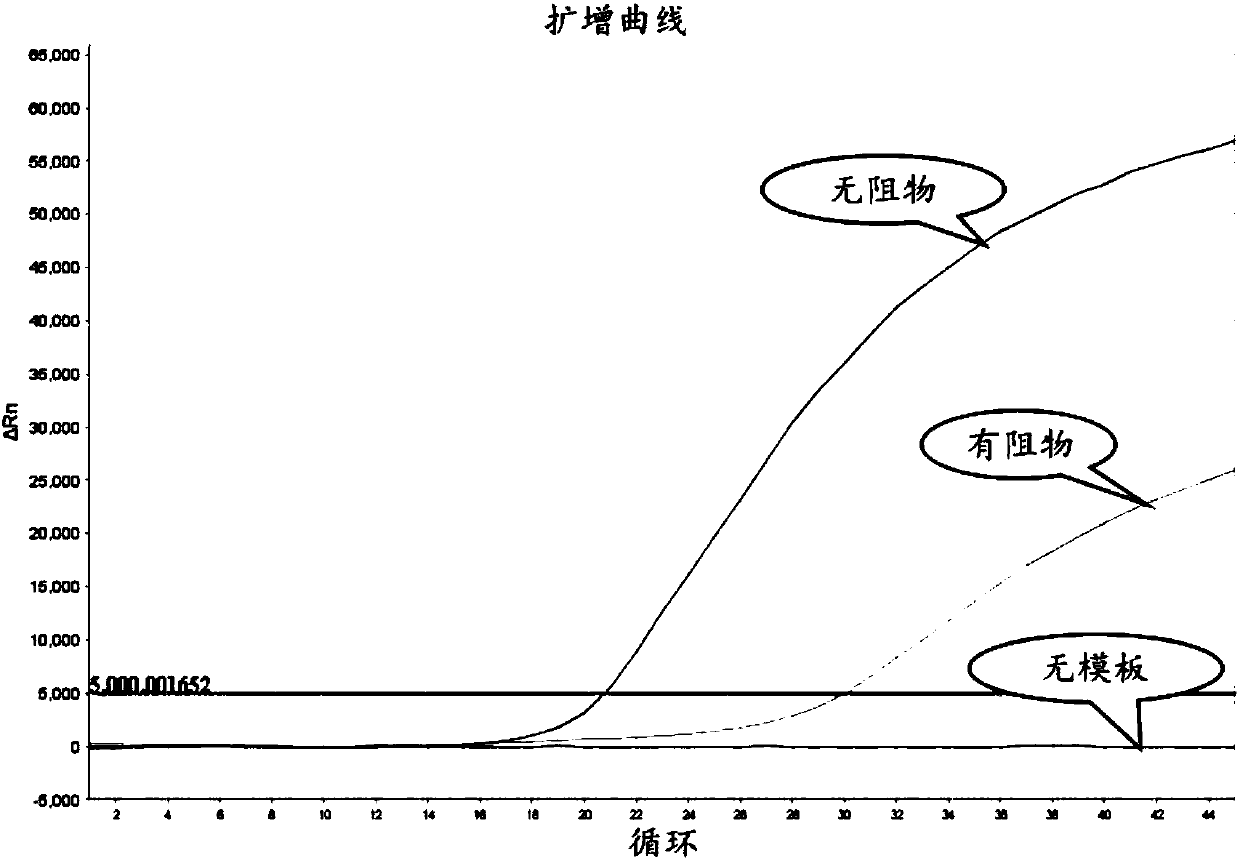
[0096] like figure 1 As shown, design the corresponding upstream forward primer, downstream reverse primer and resistance covering the mutation site according to the mutation site of the target gene to be detected. The upstream and downstream primers have no selectivity for wild type and mutant type. The blocker is designed for the wild type and has a mismatch with the mutant template. The 3'-end of the upstream primer overlaps with the 5'-end of the blocker by 2 to 6 nucleotides (nt), and the 3'-end of the blocker is blocked by PO. 4 group closed. like figure 2As shown, there is only one annealing temperature (54°C) for the primer in the first round of PCR, which is 1-5°C lower than the melting temperature (Tm-Po) of the forward primer, and the annealing time is 20s; Two annealing temperatures, one was 58°C (time 30s), for resister annealing, and the other was 54°C (time 20s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com