Method and device for identifying repetitive regions in deoxyribonucleic acid (DNA) sequences
A DNA sequence and identification method technology, applied in the field of systems biology, can solve the problems of many candidate modes, long running time, and difficulty in finding repeating sequences of DNA sequences, and achieve the effect of improving the recognition efficiency and the recognition efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0047] The specific implementation manners of the present invention will be further described in detail below in conjunction with the accompanying drawings and embodiments. The following examples are used to illustrate the present invention, but are not intended to limit the scope of the present invention.
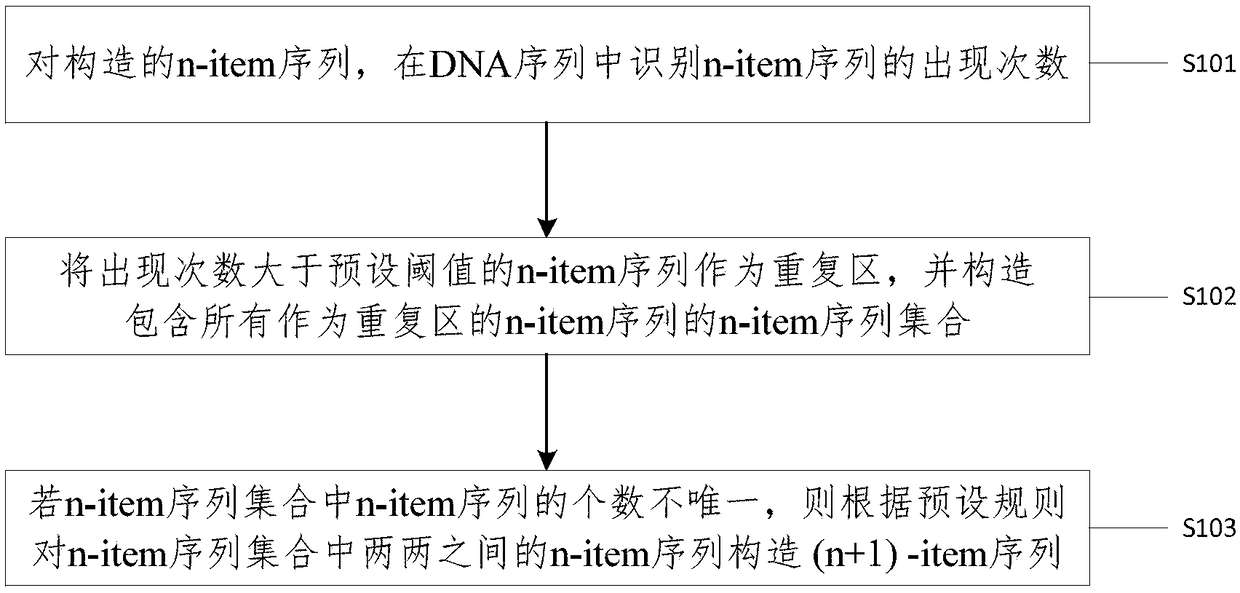
[0048] In order to overcome the above-mentioned problems in the prior art, an embodiment of the present invention provides a method for identifying repetitive regions in DNA sequences, figure 1 It is a schematic flowchart of a method for identifying repetitive regions in a DNA sequence according to an embodiment of the present invention, such as figure 1 As shown, the method includes:
[0049] S101. For the constructed n-item sequence, identify the number of occurrences of the n-item sequence in the DNA sequence.
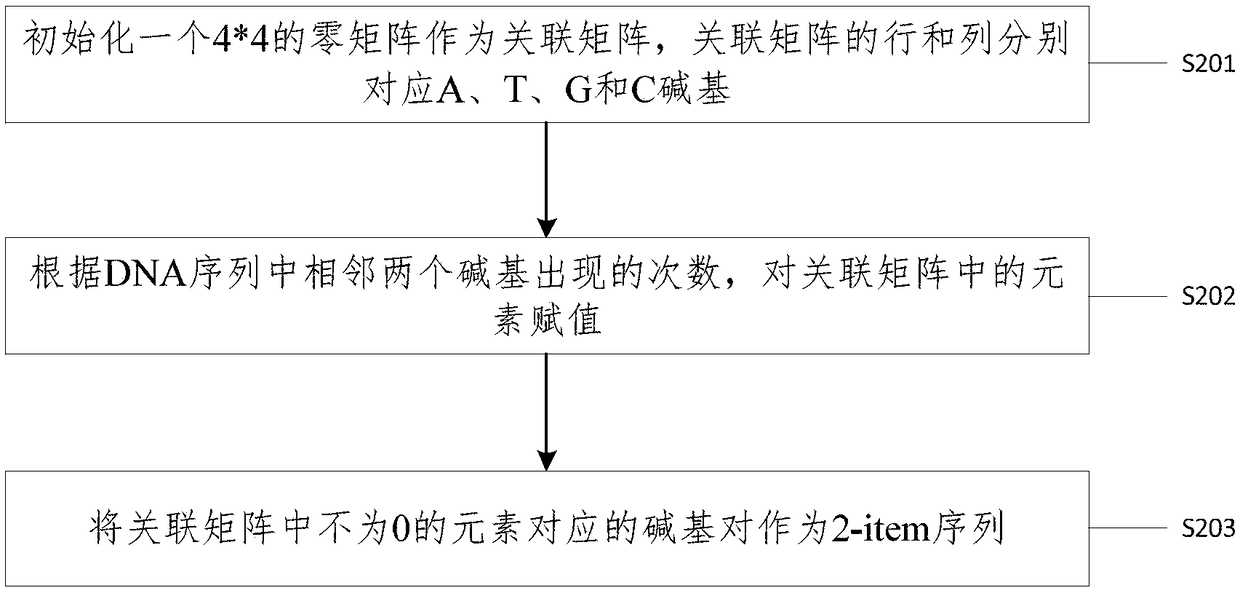
[0050] It should be noted that the n-item sequence in the embodiment of the present invention represents a DNA subsequence with a length of n and n≥2, and a l...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com