Watermelon mosaic virus detection sensor based on t4 DNA polymerase and its assembly method
A watermelon mosaic virus, detection sensor technology, applied in microorganism-based methods, biochemical equipment and methods, and microbial determination/inspection, etc., can solve the problem that target DNA has no advantage in selectivity, hinder target detection performance, and prolong reaction time. and other problems, to achieve the effect of high sensitivity and rapid detection, high sensitivity and fast response speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
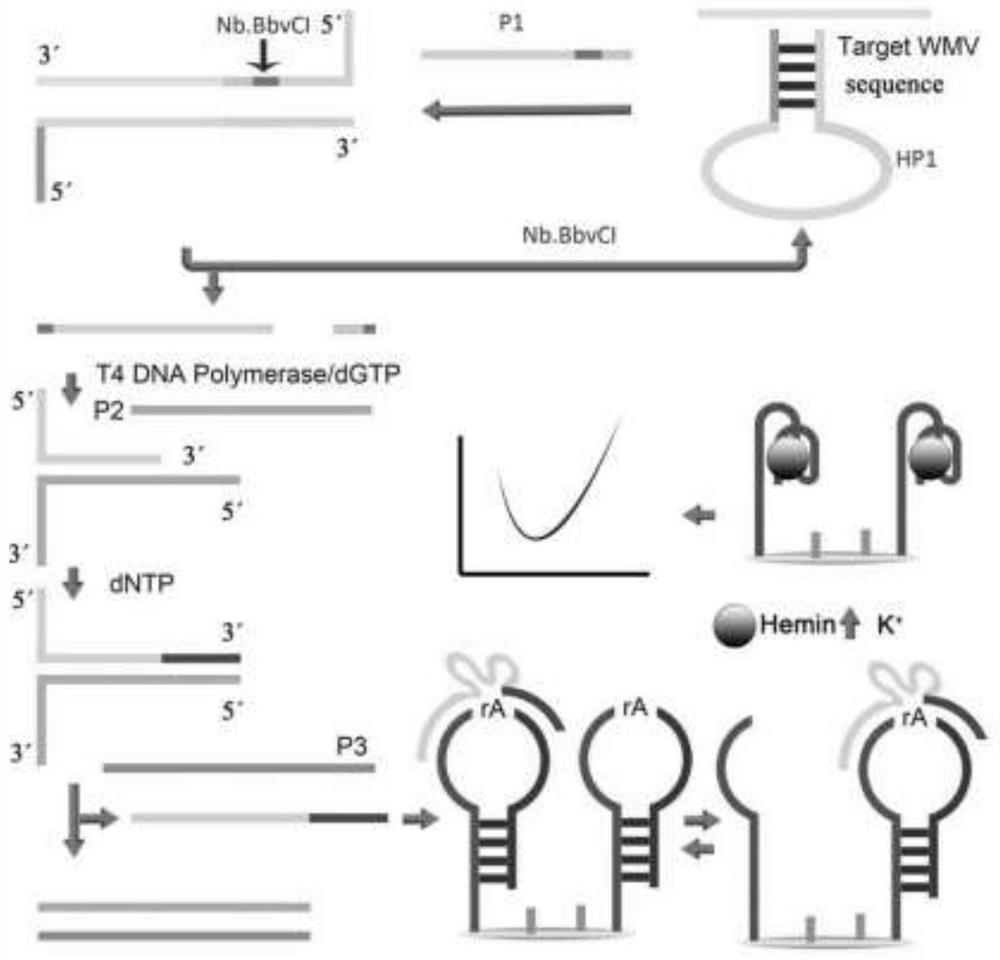
[0058] A watermelon mosaic virus detection sensor based on T4 DNA polymerase, including: probe P1, probe P2, probe P3, hairpin probe HP1, Nb.BbvCl, T4 DNA polymerase, dGTP, dNTP, hairpin probe Needle HP2, gold electrode;
[0059] Each DNA sequence is as follows:
[0060] The gene sequence of the probe P1 is shown in SEQ ID No.1:
[0061] SEQ ID No.1: 5'-ACACACAGCGATCACCCATGCCTCAGCTTTT-3';
[0062] The gene sequence of the probe P2 is shown in SEQ ID No.2:
[0063] SEQ ID No.2: 5'-GGGTTTAACATGGGTGATCGAAATAGTGGGTG-3';
[0064] The gene sequence of the probe P3 is shown in SEQ ID No.3:
[0065] SEQ ID No.3: 5'-CACCCACTATTTCGATCACCCATGTTAAACCC-3';
[0066] The gene sequence of the hairpin probe HP1 is shown in SEQ ID No.4:
[0067] SEQ ID No.4:
[0068] 5'-ATGCCTCAGATAAGCCACAAAAAAAAGCTGAGGCAT-3';
[0069] The gene sequence of the hairpin probe HP2 is shown in SEQ ID No.5:
[0070] SEQ ID No.5:
[0071] 5'-SH5'-SH-(CH2)6-TTTTGGGTTGGGCGGGATGGGTTTATrAGGTGTGTATCCCGCCC-3'.
...
Embodiment 2
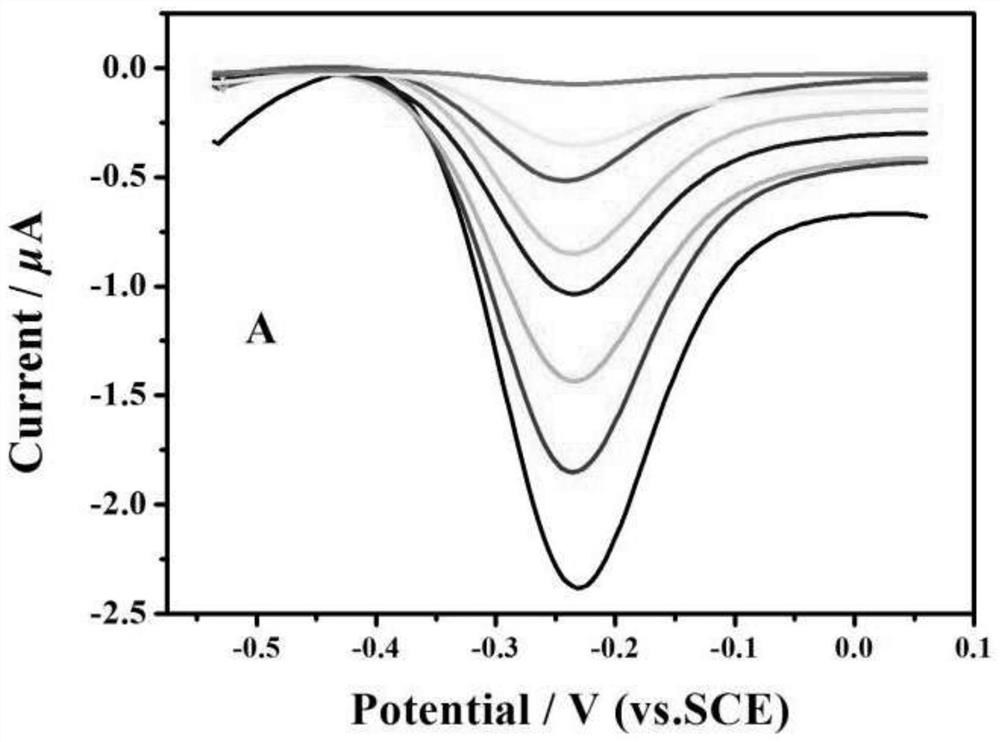
[0086] With the gold electrode obtained at last in embodiment 1 as the working electrode, the Pt electrode as the counter electrode, and the SCE electrode as the reference electrode, the current is detected by differential pulse voltammetry, and the i-t curve is recorded; the result is as follows figure 2 As shown, there is a positive correlation between the magnitude of the current value and the concentration of the target DNA (WMV cDNA).
Embodiment 3
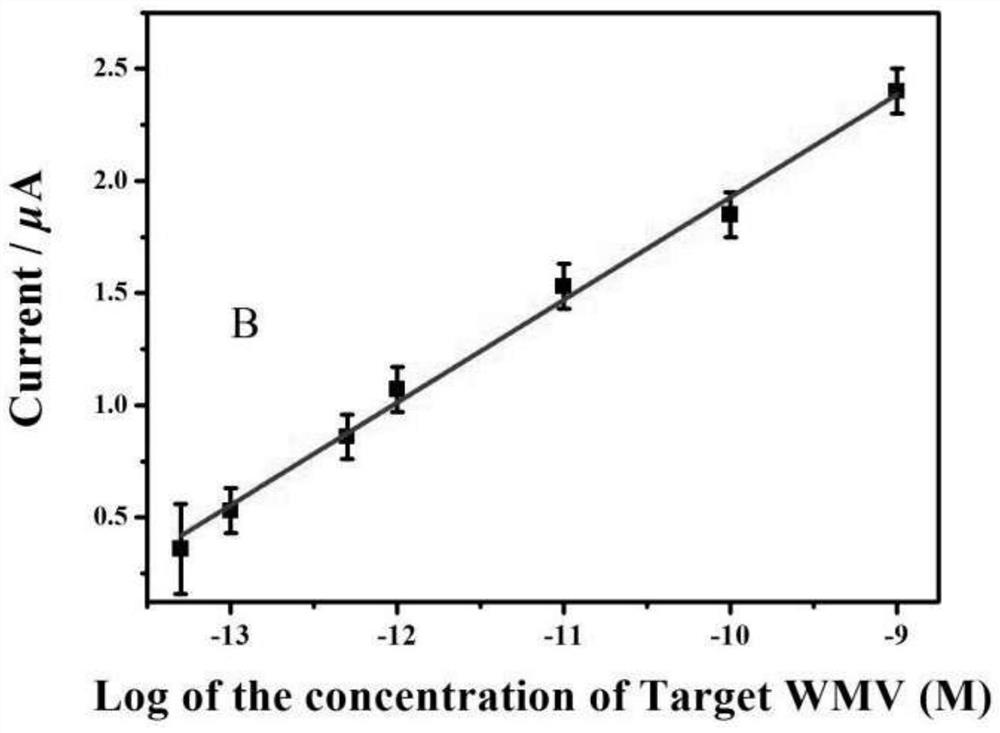
[0088] According to the current value of different concentration target DNA (WMV cDNA) solutions in implementation 1, the ordinate is taken as the abscissa with the corresponding target DNA solution concentration, and the concentration standard curve is established; the results are as follows image 3 As shown, a straight line is obtained by linearly fitting the current value and the concentration of the target DNA solution, and the linear correlation is above 0.99. The detection limit is calculated by the 3σ method, and the linear range is 50fM-1nM, and the minimum detection limit is 50fM.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com