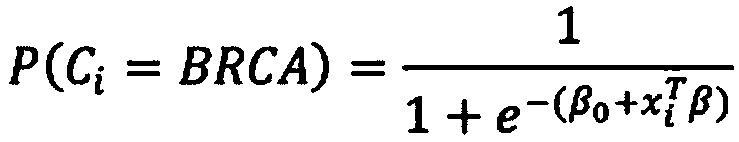Method of characterising a DNA sample
A sample and characterization technology, applied in biochemical equipment and methods, microbial determination/inspection, instruments, etc., can solve the problems of tumor cell apoptosis, lack of effective repair of double-strand breaks, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
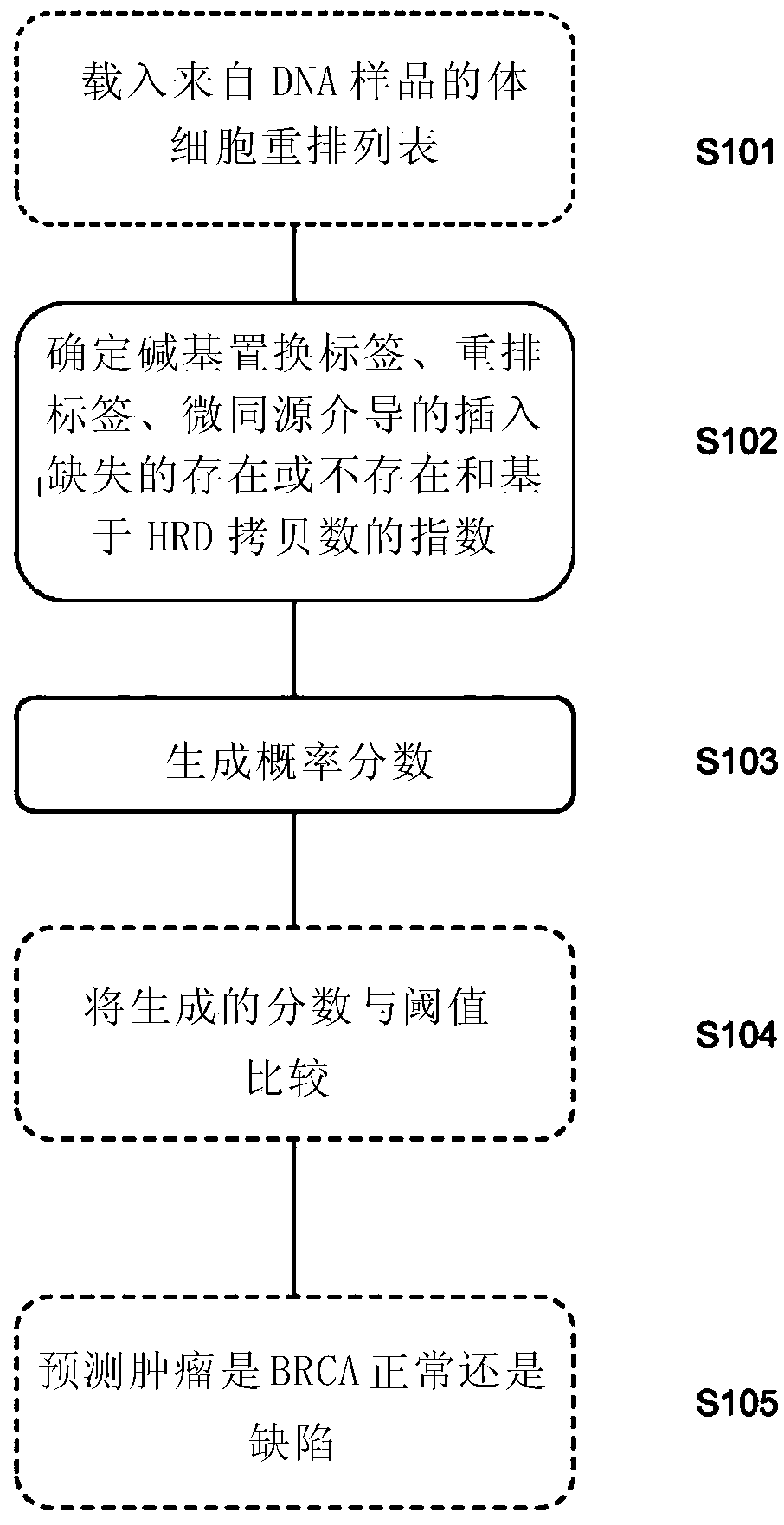
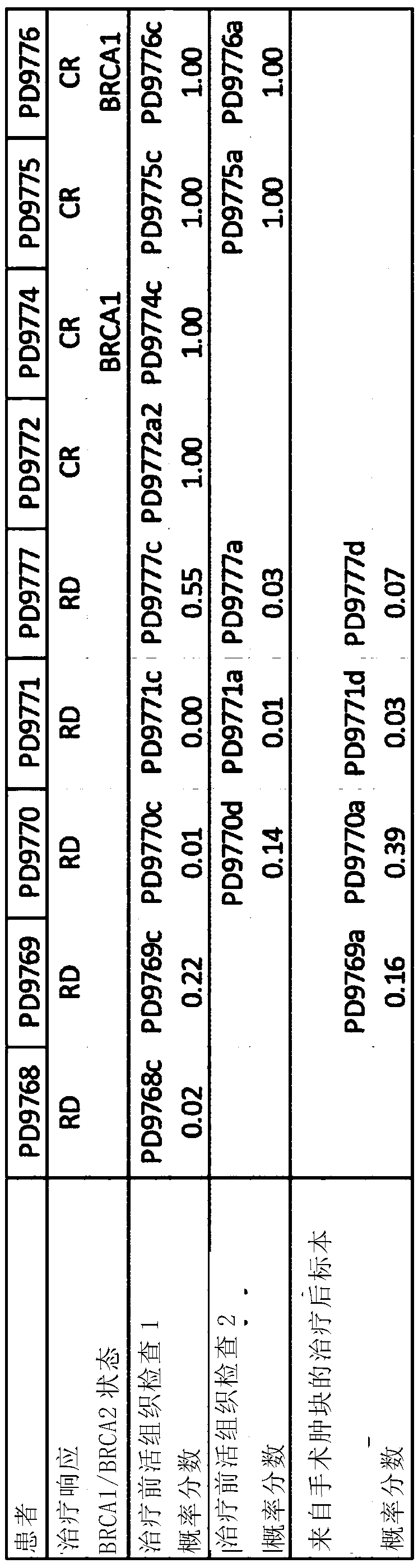
[0017] A first aspect of the invention provides a method of characterizing a DNA sample from a tumor comprising the steps of: determining the presence or absence of a plurality of base substitution tags, rearrangement tags and one or more indels in the sample tags, and a copy number profile of the sample; generating a probability score based on the presence or absence of said plurality of base substitution tags, rearrangement tags, and indel tags in the sample, and the copy number profile of the sample; and Score, identifying whether the sample has a high or low likelihood of being homologous recombination (HR) deficient.
[0018] A second aspect of the invention provides a method of characterizing a DNA sample from a tumor comprising the steps of:
[0019] Do two or more of the following steps:
[0020] a) Determining the presence or absence of at least one base substitution tag in the sample
[0021] b) determining the presence or absence of at least one rearranged tag in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com