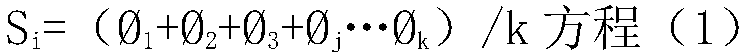Biological sample intelligent identification method based on molecular map
A biological sample and intelligent identification technology, applied in the field of intelligent identification of biological samples based on molecular maps, can solve the problems of lack, difficulty in meeting the classification and identification requirements of large biological samples, and excess features, so as to achieve fast analysis speed, improve feasibility and Efficiency, increased accuracy and speed effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
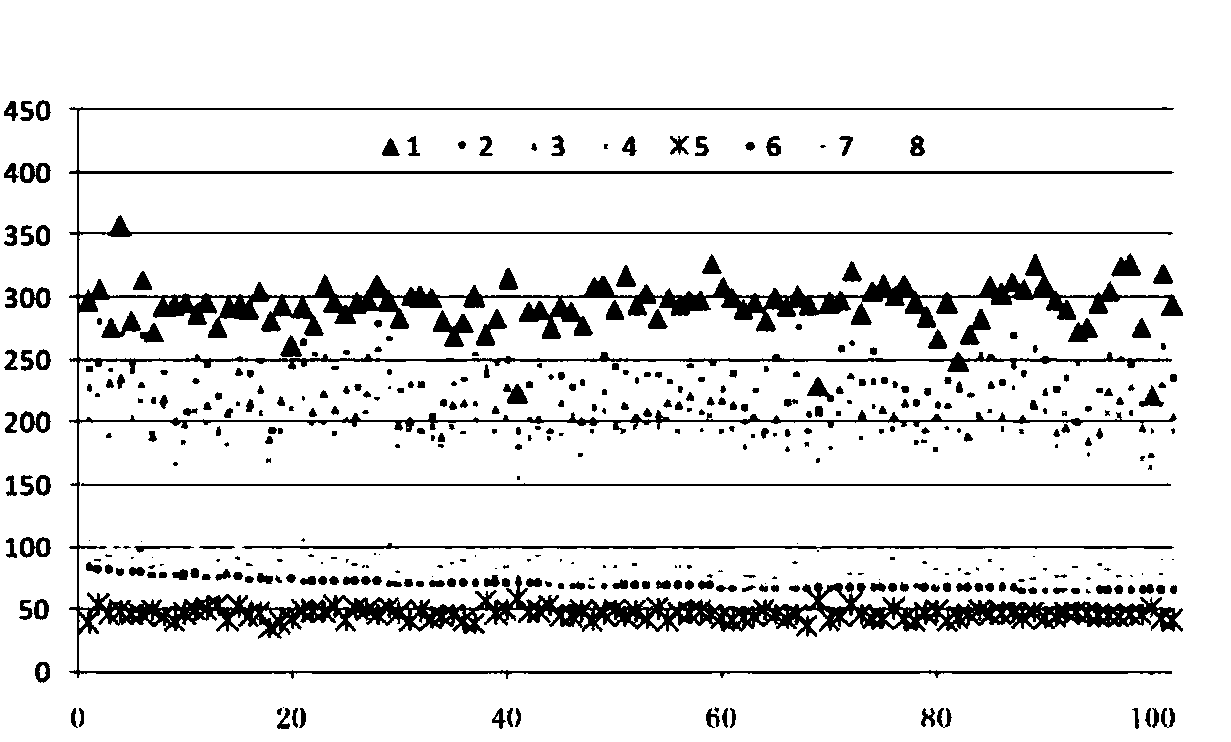
[0059] The following example demonstrates how to use molecular map-based point cluster matching technology to identify and classify ginseng, American ginseng, red ginseng, panax notoginseng, achyranthes bidentata, rehmannia glutinosa, sophora flavescens, and astragalus with high precision.
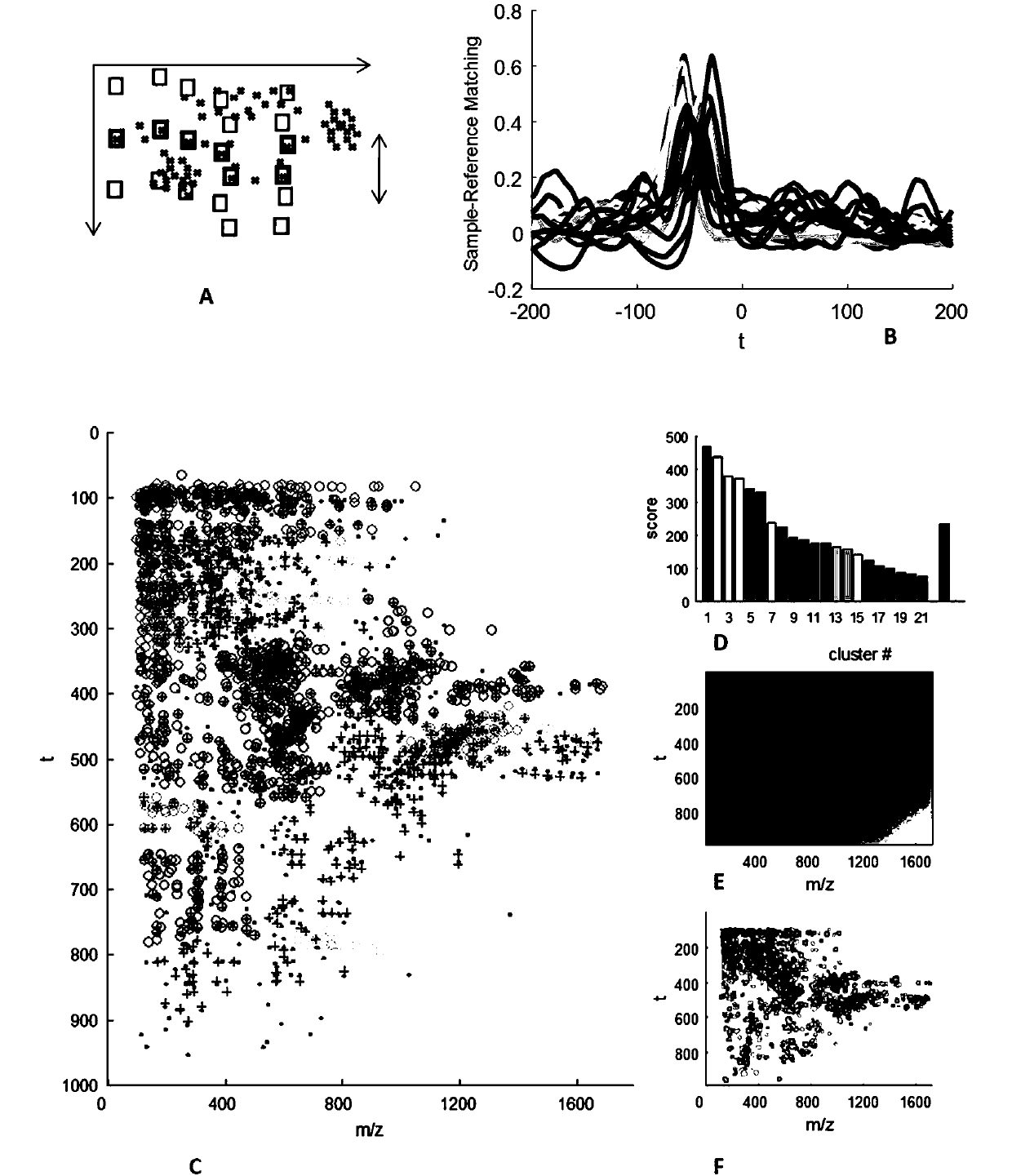
[0060] The purpose of this example is to demonstrate how to use molecular map generation technology, point density-based clustering technology, image segmentation technology, cluster matching technology, machine classification technology, etc. to achieve efficient and accurate identification of samples.
[0061] Materials and Methods:
[0062] Chinese medicine samples from the market: ginseng (1; number of samples n=170), American ginseng (2; number of samples n=100), red ginseng (3; number of samples n=100), Panax notoginseng (4; number of samples n=100 ), Achyranthes bidentata (5; sample number n=100), Rehmannia glutinosa (6; sample number n=100), Sophora flavescens (9; sample number n=1...
Embodiment 2
[0091] The following example demonstrates how to use molecular map-based point cluster matching technology to identify and classify Albizia juliensis and Albizia juliensis with high precision.
[0092] The purpose of this example is to demonstrate how to use molecular map generation technology, point density-based clustering technology, image segmentation technology, cluster matching technology, machine classification technology, etc. to achieve efficient and accurate identification of samples.
[0093] Materials and Methods:
[0094] U, Chinese medicine samples from the market Albizia Julibrissin (n=100), Albizia Julibrissin (n=100) as unknown samples (sample to be tested); =52), Sophora flavescens (9; n=192), Astragalus membranaceus (10; n=212) as training samples, and another 10 kinds of medicinal materials (ginseng, American ginseng, red ginseng, Panax notoginseng, Achyranthes bidentata, Rehmannia glutinosa, Acacia bark, Albizia julibrissin, Sophora flavescens, Astragalus...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com