Closely linked molecular marker of wheat spike grain number main effect QTL and application of closely linked molecular marker
A technology of molecular markers and traits of grain number per ear, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., to achieve the effects of saving production costs, fast and accurate screening, and clear selection targets
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Example Embodiment
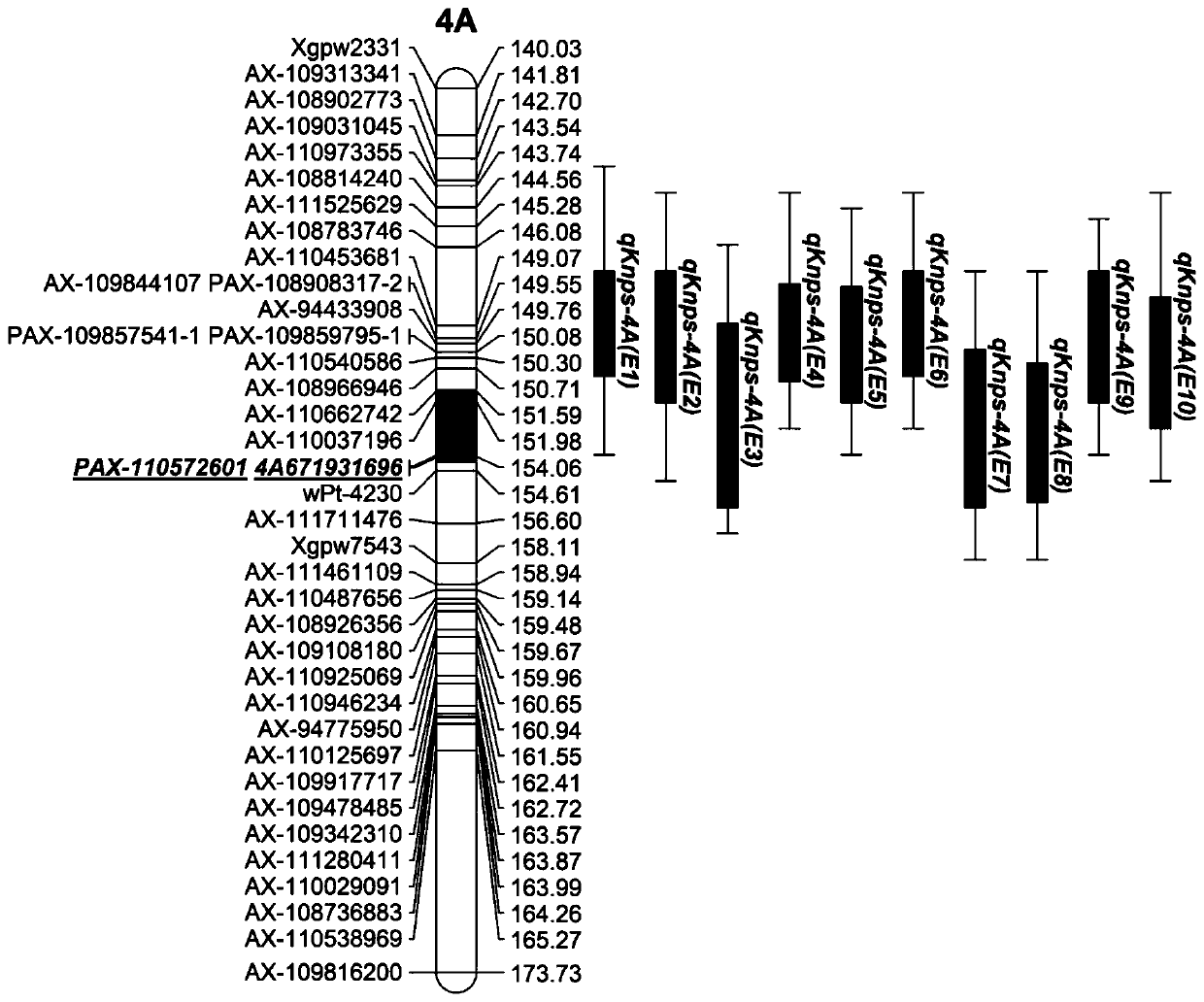
[0036] Example 1 Molecular markers closely linked to the major QTL for wheat spike number
[0037] Use 4A671931696 labeled primer:
[0038] Upstream primer sequence: GCTGGGGTTTCTCTGCAAAA (as described in SEQ ID NO:1),
[0039] Downstream primer sequence: TGGGGAGTGGCATCTTGTAG (as described in SEQ ID NO: 2),
[0040] Use PAX-110572601 labeled primers:
[0041] Upstream primer L 1 Sequence: TTATCTACTACCCATTTCCACCCTAAC (as described in SEQ ID NO: 3),
[0042] Upstream primer L 2 Sequence: TACTACCCATTTCCACCCCCAT (as described in SEQ ID NO: 4),
[0043] Downstream primer R sequence: TGGTTGTAATCAGTCCATGT (as described in SEQ ID NO: 5);
[0044] The 4A671931696 labeled primer was used for PCR amplification of the DNA of wheat variety Jing 411. The PCR amplification system was 10 μl, including: 1 μl DNA template, 1 μl upstream primer, 1 μl downstream primer, 5 μl 2×Taq PCR StarMix, 2 μl DdH 2 O; Amplification by ordinary amplification program: denaturation at 95°C for 5 min; 34 cycles of ordinary ...
Example Embodiment
[0048] Example 2 Screening method and application of molecular markers closely linked to the main QTL for wheat spike number
[0049] It includes the following steps:
[0050] (i) Crossing the wheat variety Jing 411 with more grains per spike as the female parent and Kenong 9204 with fewer grains per spike as the male parent to obtain hybrid F1, F2 produced by F1 selfing, and F2 selfed generation by generation F6 generation RIL population with 188 families;
[0051] (ii) Use a modified CTAB method, that is, a modified cetyltrimethylammonium bromide method (Vander Beek et al., 1992) to extract DNA from each strain of the above-mentioned RIL population, using InDel labeling, based on expressed sequence tags PCR amplification marker (STS marker) performs genotype analysis on the family to obtain the genotype value material of the RIL population ( figure 1 ); sequence tag site (sequence tag site, STS), also known as sequence target site, uses primers for PCR amplification to combine wit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com