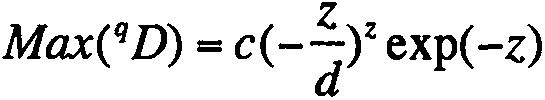Method for predicting microflora structure based on power law scaling model of diversity-area relationship (DAR)
A microbial community and diversity technology, applied in the fields of computational biology and medical ecology, can solve problems such as the inability to fully reflect the distribution of microbial communities, and achieve the effect of reliable technical indicators
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1: Monitoring the structure and distribution of intestinal flora in 196 obese patients
[0050] Step【1】Citation of the data source: The gut microbiota site data were collected from 196 obese patients (BMI≥30kg m -2 ), the intestinal flora of obese patients constituted a special microbial ecosystem, and each patient’s fecal flora sample was a site sample in the microbial ecosystem, a total of 196 site samples. The original author used 16s-rRNA gene marker sequencing technology to obtain data on the species richness (Richness) and abundance (Abundance) of intestinal microbial flora in obese people.
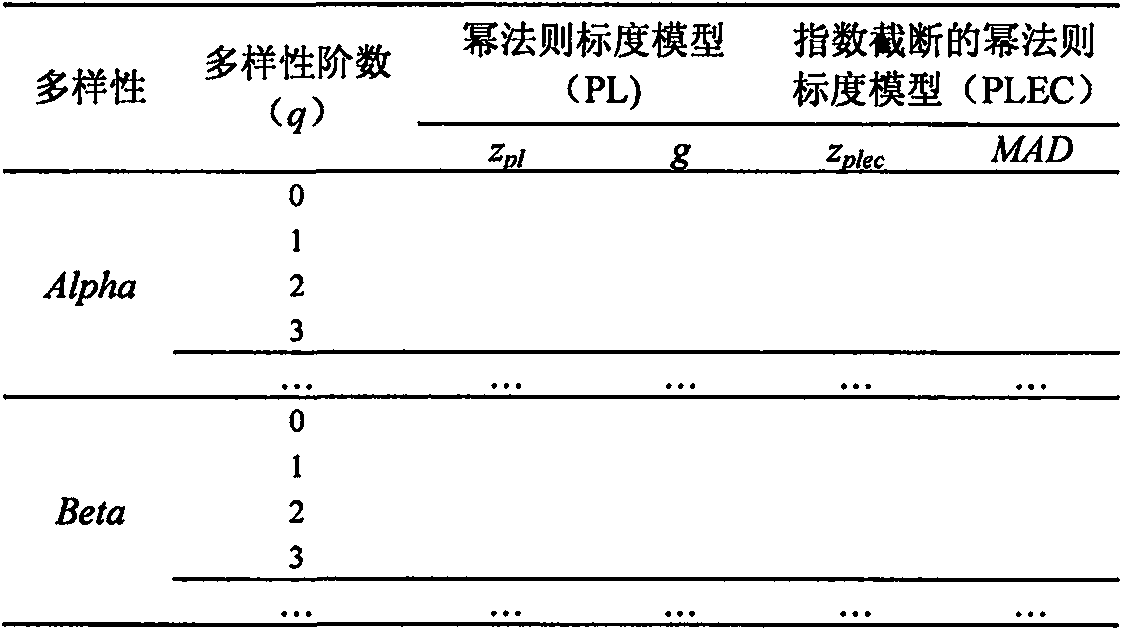
[0051] Step [2] Data processing: The 196 sampling sites of the intestinal microbial flora data of obese patients were randomly sorted, and the same number of sample sequences were randomly selected from these equally probable random sequences. The representative data can be computed to fit a DAR model.
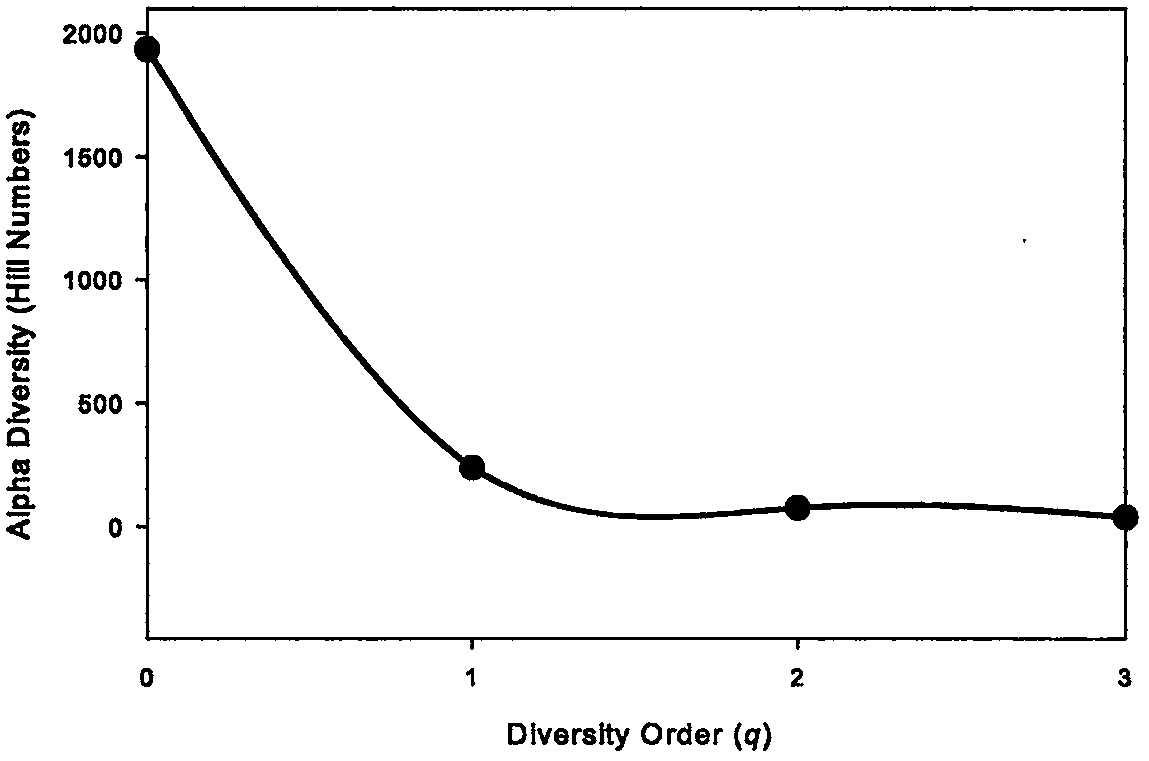
[0052] Step [3] Calculate the diversity index (Hill numbers): ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com