Peptide fragment-spectrogram matching credibility testing method and system, storage medium and device
A test method and reliability technology, applied in the field of computational proteomics, which can solve problems such as insufficient quality control of identification results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
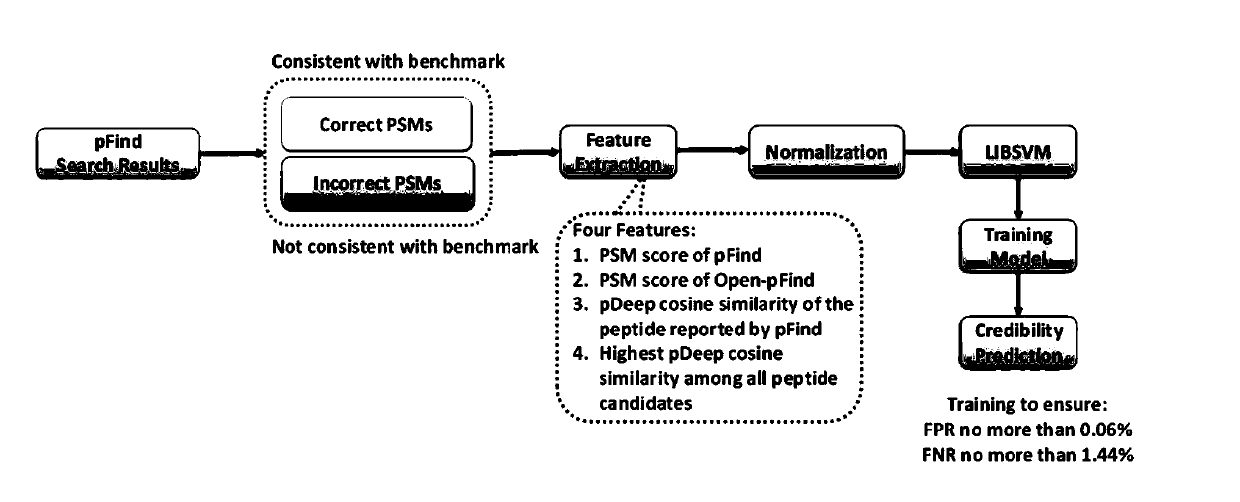
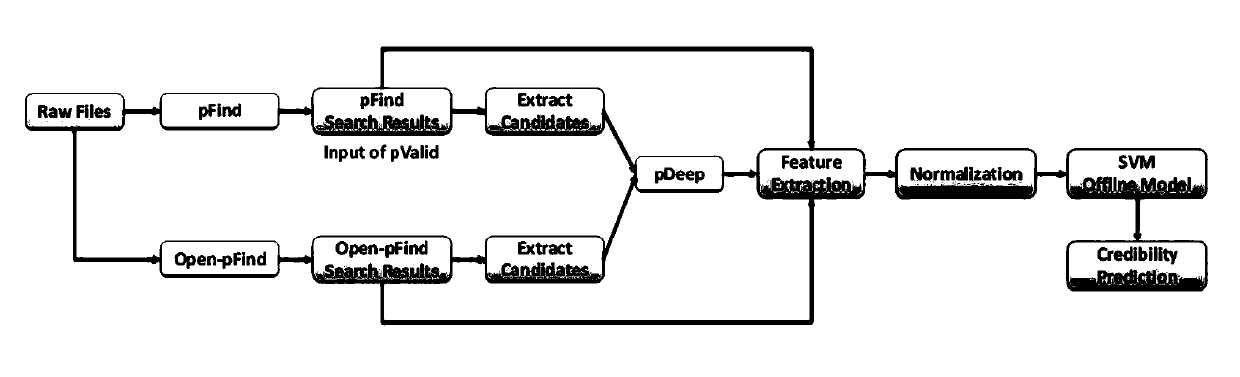
[0054] The credibility testing method proposed by the present invention is just to solve the problems existing in the above-mentioned credibility testing method. The present invention solves the following three technical problems: 1) individual credibility testing is carried out on the identification results; 2) guarantee testing Accuracy of results; 3) Rapid and efficient automated inspection of large-scale identification results.
[0055] At the problems referred to above, the present invention proposes following key points:
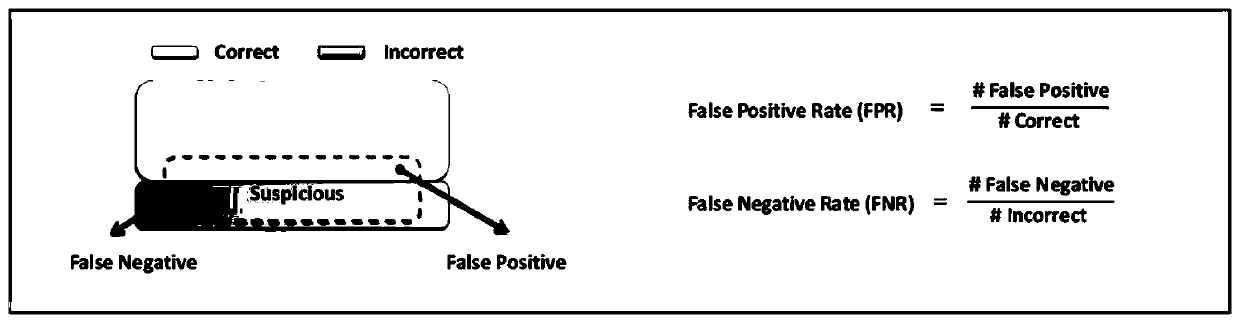
[0056] Key point 1, two evaluation indicators for the reliability test method are proposed - FPR (False Positive Rate, False Positive Rate) and FNR (False Negative Rate, False Negative Rate). FPR measures the proportion of a reliability test method that discriminates true and correct identification results as doubtful identification results, and FNR measures the proportion of this reliability test method that discriminates true and false identification...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com