Solid tumor polygene detection gene chip and preparation method and detection device thereof
A technology of gene chip and detection method, applied in the field of tumor detection, can solve the problems of long cycle, large sample consumption and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0111] The technical solution of the present invention is compared with the Chinese invention patent titled "gene chip for detection of tumor mutation burden and its preparation method and device" and the application number is 2018107129399. The main differences include:
[0112] a) Delete the genes shown in Table 5 below. These genes have a low contribution to the calculation of tumor mutation burden and are not genes related to targeted drugs.
[0113] table 5
[0114]
[0115]
[0116] b) Add the genes shown in Table 6 below. These genes have a high contribution to the calculation of tumor mutation burden, which can improve the accuracy of tumor mutation burden calculation.
[0117] Table 6
[0118] APOBEC3A LOC101927322 PRKACA TIMM23B ARTN MAP4K3 PRKDC TMEFF1 ATP2C1 METTL21A PROC TOE1 BCL2L11 MFSD11 RAD17 TPM3 C11ORF65 MYCNOS RAD50 U2AF1L5 C1QTNF5 NBPF10 REEP5 UGT1A10 CD3EAP NBPF19 RHO...
Embodiment 1
[0135] In this example, custom probes were designed, covering the probe sequences of Tables 1 to 4 and 28 SNP site regions. The chip performance test results are shown in Table 7 below. It can be seen that the probe design is compatible with the common Illumina platform and MGI platform, and the coverage of the design area reaches more than 99%, which meets the normal use requirements.
[0136] Table 7
[0137] sample number Sequencing platform Capture efficiency (%) Coverage(%) DNR901971SLZAA01_3_2 MGI 37.195 99.829 DNR901971XYZAA01_2_3 MGI 42.099 99.792 DNR901979SLZAA01_3_2 MGI 32.582 99.868 DNR901979XYZAA01_2_3 MGI 40.51 99.838 DNR902340SLZAA01_2_1 MGI 49.45 99.789 DNR902340XYZAA01_2_1 MGI 39.764 99.764 DNR903896SLZAA01_3_2 MGI 38.427 99.857 DNR903896XYZAA01_2_3 MGI 56.814 99.76 DNR904698SLZAA01_2_1 MGI 42.357 99.755 DNR904698XYZAA01_2_1 MGI 40.17 99.792 DNR900409SL...
Embodiment 2
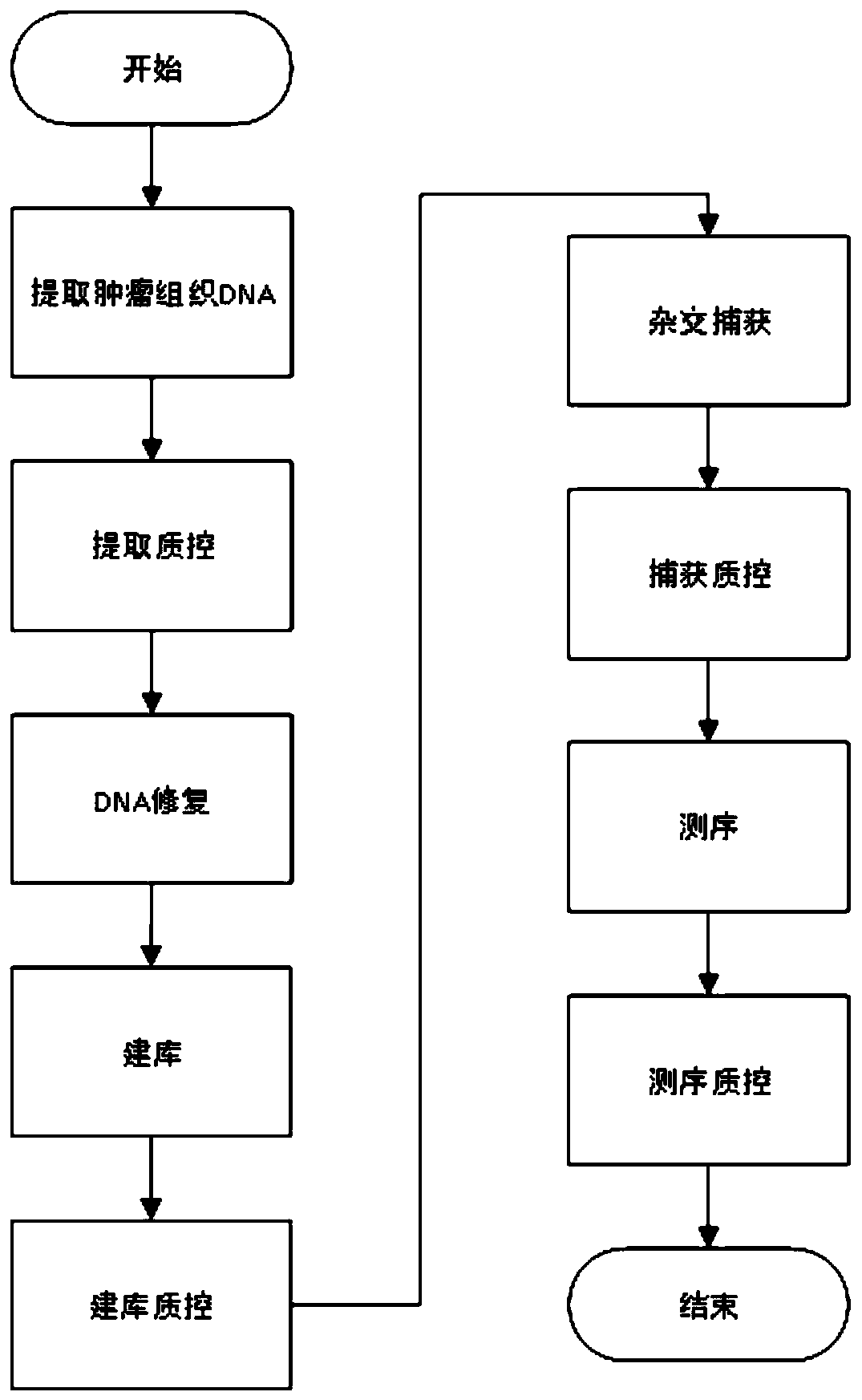
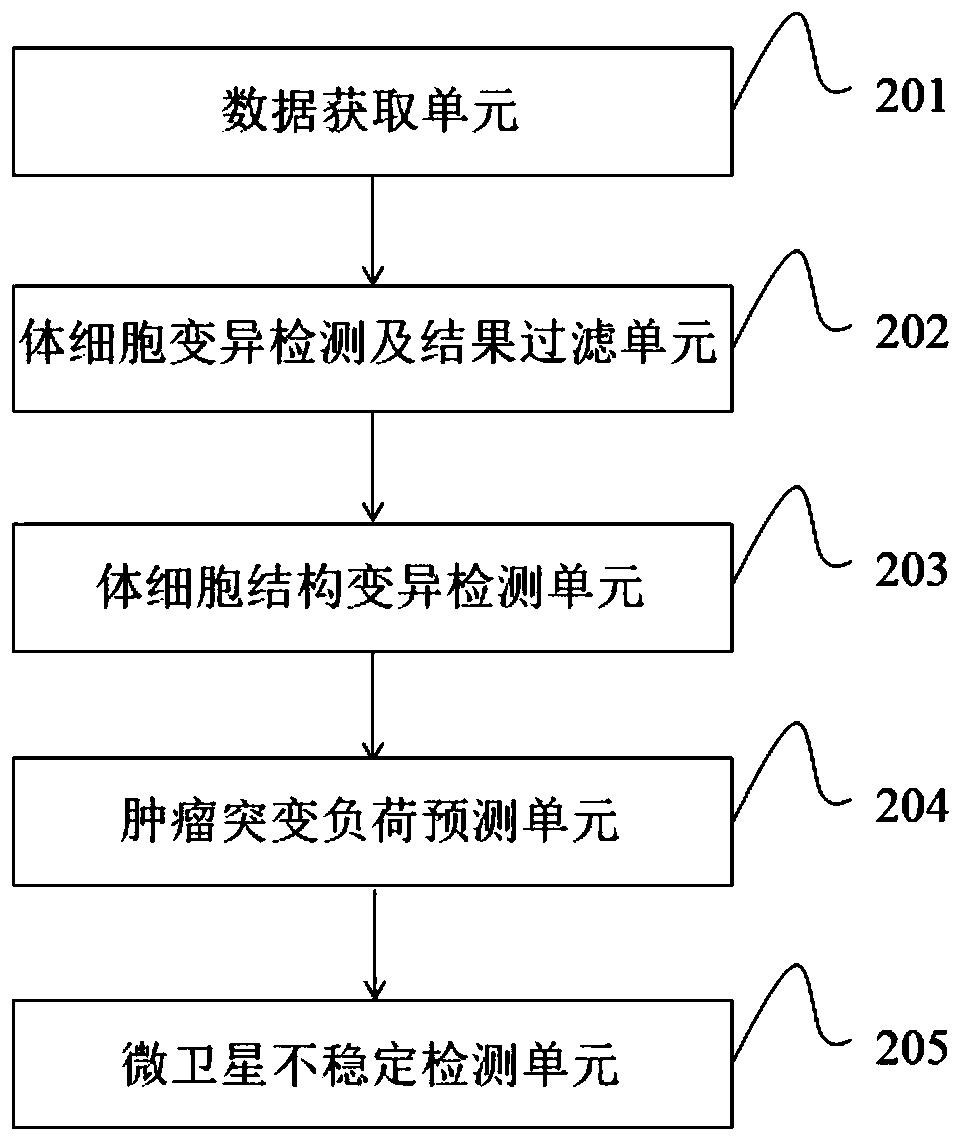
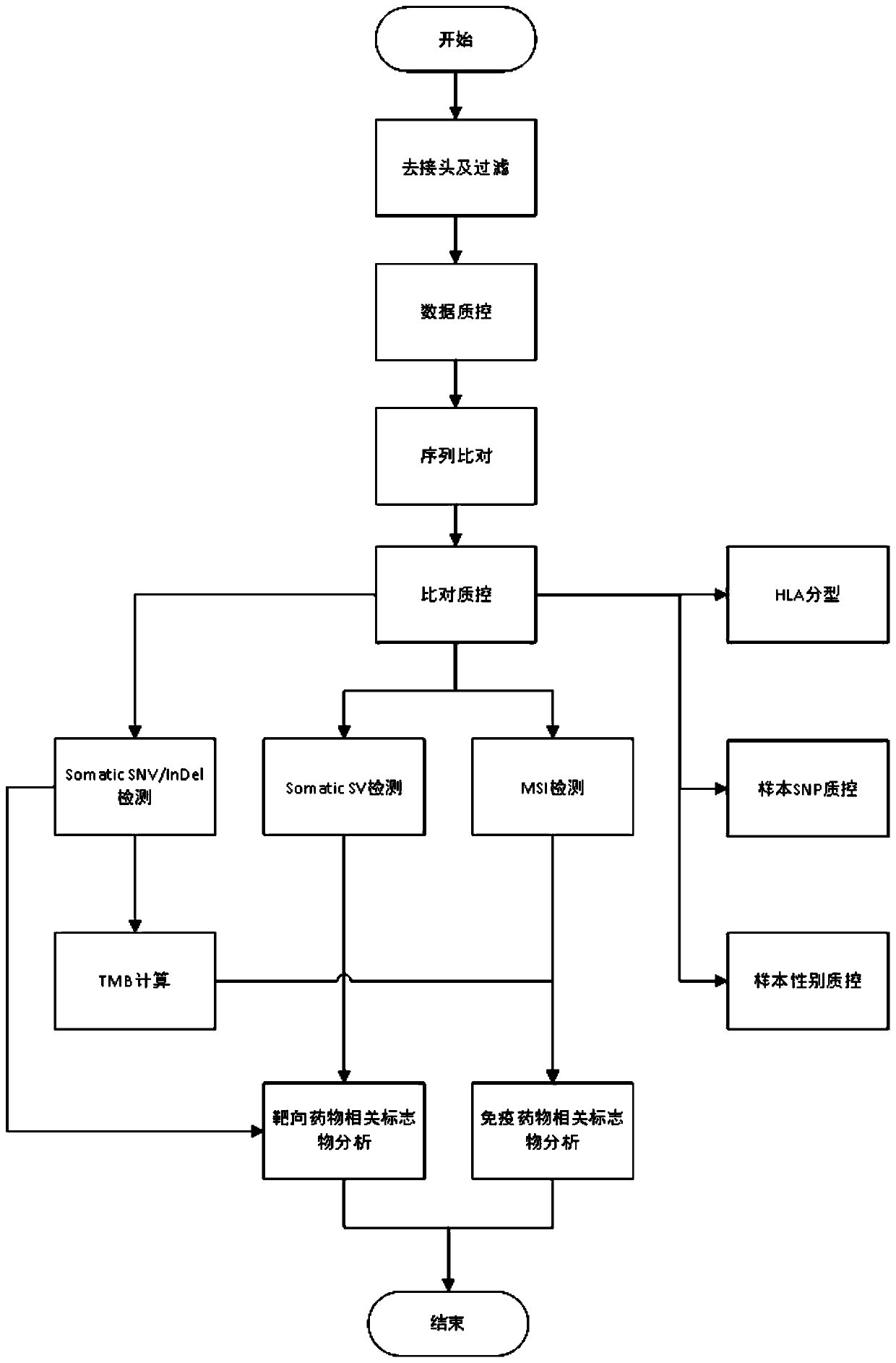
[0139] This example shows the information analysis method, including off-machine data processing, data filtering and quality control, sequence comparison and result quality control, somatic cell mutation detection and result filtering, mutation result annotation, MSI detection, tumor purity prediction, TMB prediction , TMB medication guidance and other links. The above-mentioned information analysis method runs on the bioinformatics analysis cluster through the automatic scheduling system, and outputs the analysis results stably and efficiently. Such as image 3 As shown, each information analysis link is introduced in detail as follows:
[0140]a) Off-machine data processing: The data generated by the sequencer is usually in a special format, which needs to be converted into a common fastq file format first. In addition, multiple samples will be mixed on one sequencing chip, and the data belonging to each sample needs to be split before analysis. This method can adapt to t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com