Liver cancer early warning method
A liver cancer and genetic technology, applied in the field of early warning of liver cancer, can solve the problems of high missed diagnosis rate and misdiagnosis rate, poor effect, low sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0273] Example 1 Plasma cfDNA Extraction
[0274] 1.1 Separation and storage of plasma samples
[0275] Take 8mL of peripheral blood from the subject and centrifuge at 2500rpm at 4°C for 15min to separate the sample into three layers: plasma, white blood cells and red blood cells. Use a pipette gun to take out the uppermost layer of plasma in the anticoagulant tube and divide it into several 1.5mL centrifuge tubes After labeling, store at -80°C.
[0276] 1.2 cfDNA extraction
[0277] A commercial plasma cell-free DNA extraction kit (Guangzhou Meiji Biotechnology Co., Ltd., D3182-03A) was used, and all operations were strictly performed in accordance with the kit instructions.
[0278] 1.3 cfDNA quality detection
[0279] BioAnalyzer 2100 (Agilent) was used to detect the quality of the extracted cfDNA. When the test results showed that the main peak of cfDNA was distributed around 160bp, it indicated that the quality of the extracted cfDNA met the requirements.
[0280] 1.4...
Embodiment 2
[0293] The design of embodiment 2 primers and probes
[0294] According to the sequence of the determined target gene, a series of specific primers and detection probe sequences as shown in Table 2 were designed. The sequences of the primers and probes used in this experiment are respectively: SEQ ID No: 21 as the upstream primer of the P16 gene, SEQ ID No: 22 as the downstream primer of the P16 gene, and SEQ ID No: 23 as the probe of the P16 gene; SEQ ID No:25 is used as the upstream primer of SFRP1 gene, and SEQ ID No:26 is used as the downstream primer of SFRP1 gene, and SEQ ID No:27 is used as the probe of SFRP1 gene; SEQ ID No:36 is used as the upstream primer of RASSF1A gene, SEQ ID No:37 is used as the downstream primer of RASSF1A gene, and SEQ ID No:38 is used as the probe of RASSF1A gene; SEQ ID No:44 is used as the probe of GSTP1 gene, and SEQ ID No:42 is used as the upstream primer of GSTP1 gene, SEQ ID No: 43 is used as the downstream primer of GSTP1 gene; SEQ ID ...
Embodiment 3
[0299] Embodiment 3 detection system research
[0300] 3.1 Determine the PCR reaction conditions
[0301] The test results showed that the P16, SFRP1, RASSF1A, and β-actin quadruple gene detection system had high sensitivity and specificity under the PCR reaction conditions shown in Table 3, and the GSTP1, APC, β-actin triple gene detection system was in The PCR reaction conditions shown in Table 4 have higher sensitivity and specificity, and the PCR amplification program is shown in Table 5.
[0302] table 3
[0303]
[0304]
[0305] Table 4
[0306]
[0307]
[0308] table 5
[0309]
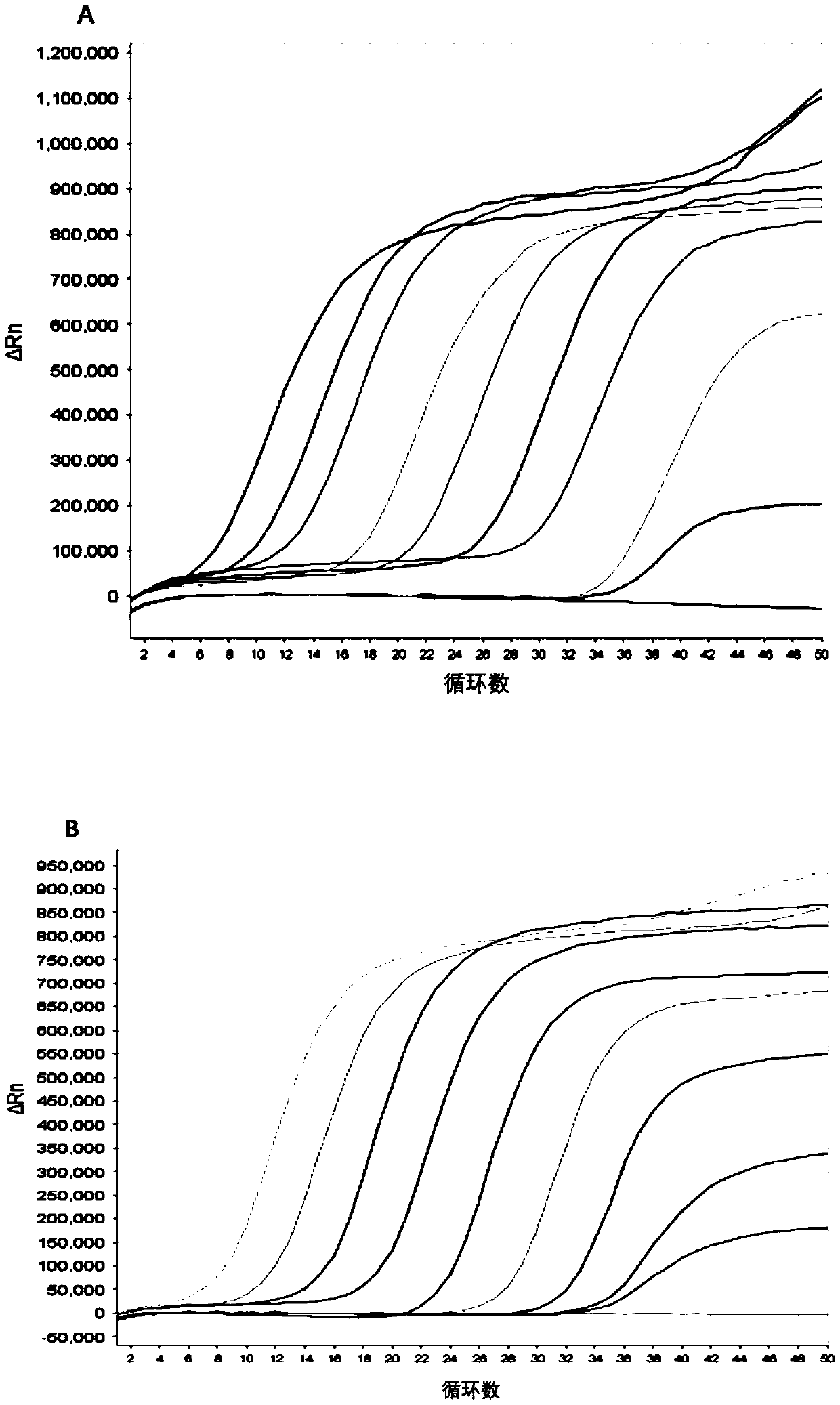
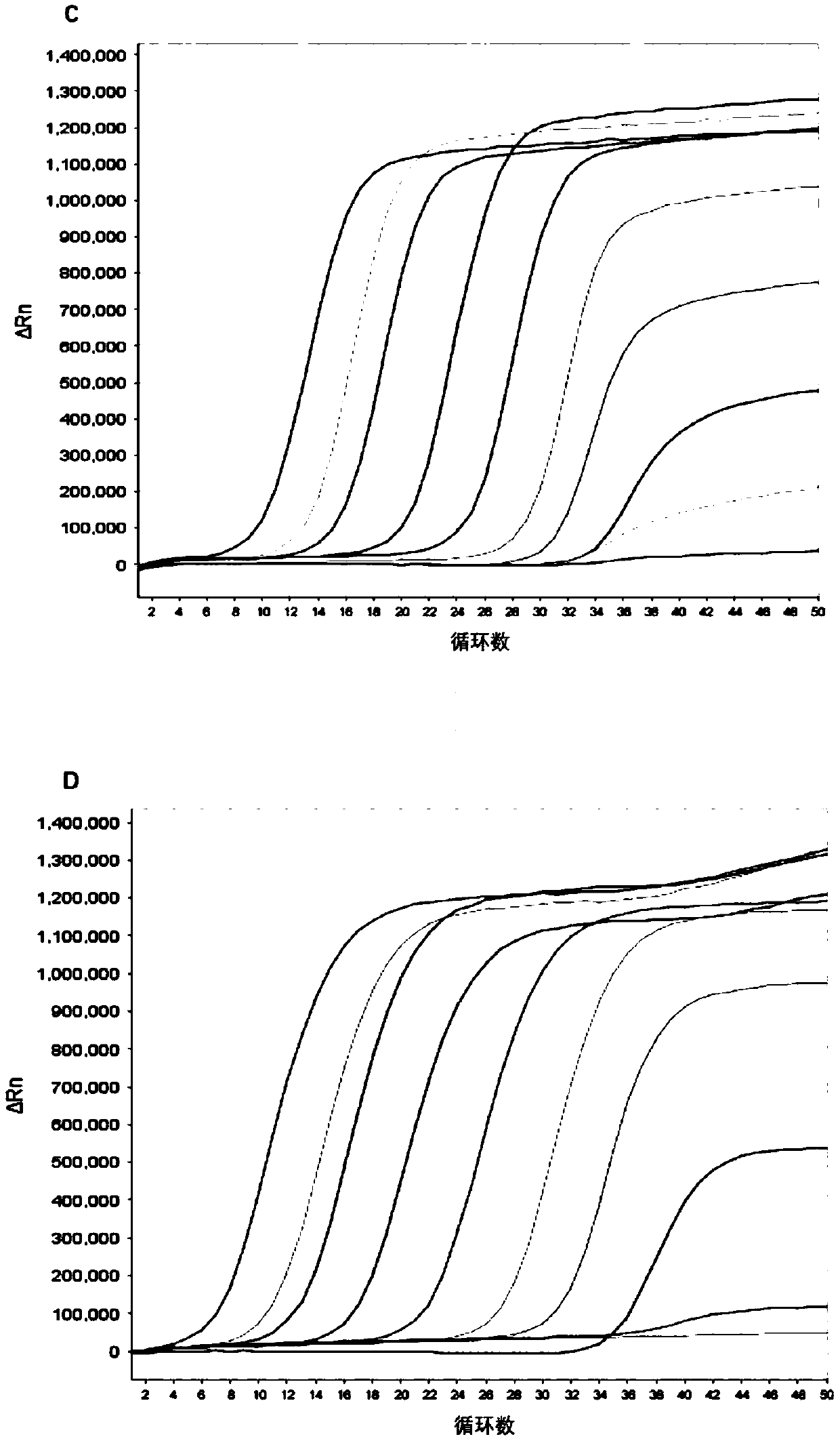
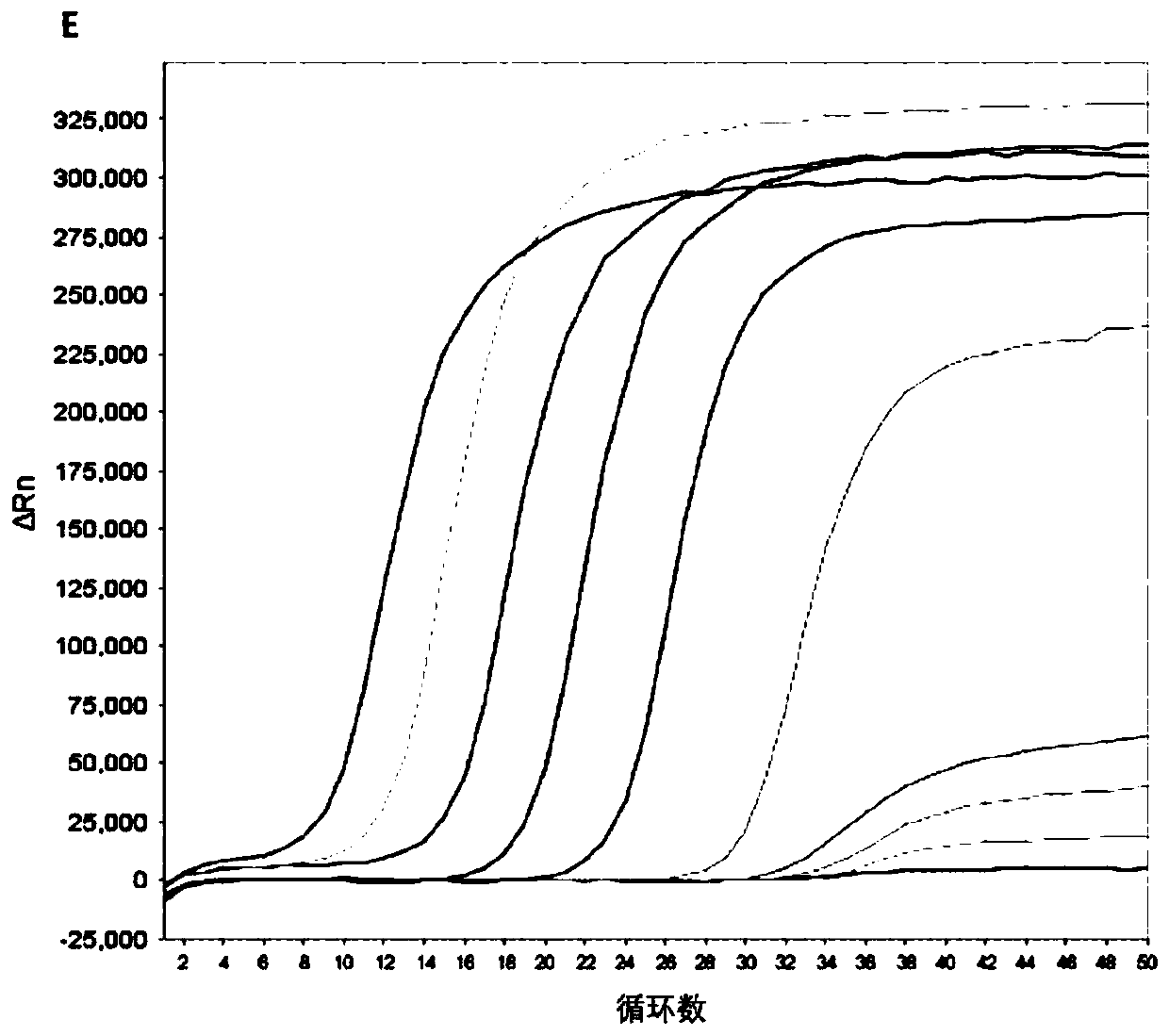
[0310] 3.2 Sensitivity Verification
[0311] Generally, the content of cfDNA in the human body is 10-15ng / mL plasma, and the half-life is 0.5-2h, so it is necessary to establish a highly sensitive detection method.
[0312] First, gene fragments were synthesized according to the positive quality control sequences of P16, SFRP1, RASSF1A, GSTP1, and APC genes in Table 1 and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com