A method for improving the efficiency of site-directed modification of cell genome by using modified ssodn
A genome-specific and site-modified technology, applied in the field of genetic engineering, can solve problems such as low efficiency of site-specific modification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Method for Improving the Efficiency of Site-directed Modification of Human Cell Genome
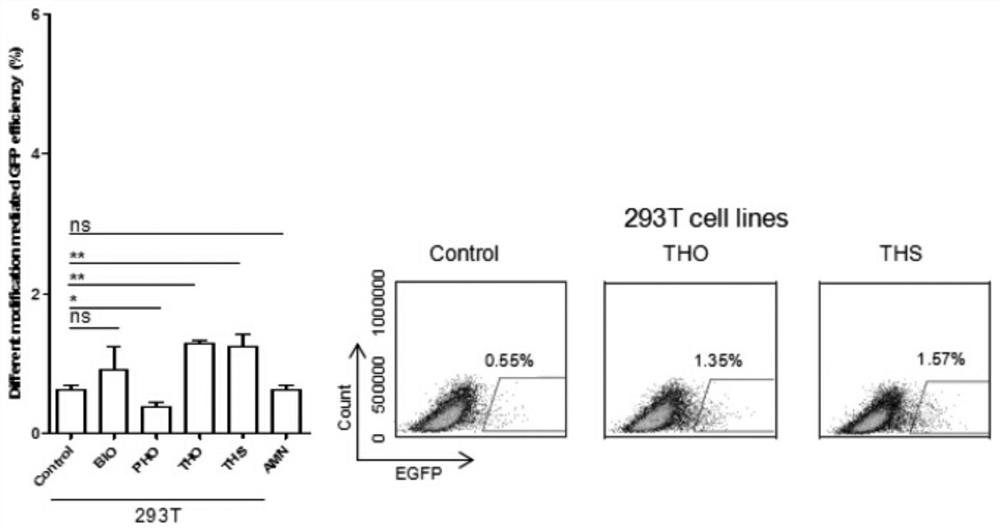
[0026] In this example, human-derived cells (293T) were used as a tool cell line for experiments.
[0027] 1. Plasmid construction
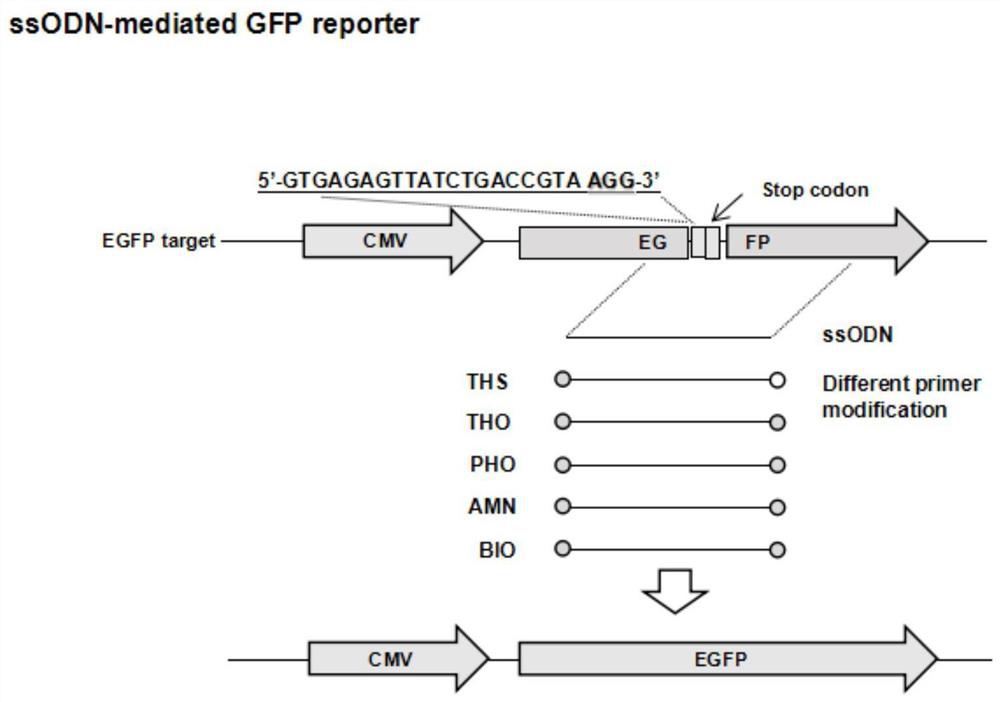
[0028] Use the online website (https: / / crispr.cos.uni-heidelberg.de / index.html) to design the pig ROSA26 gene gRNA sequence (the gene sequence is shown in SEQ ID No:1 and SEQ ID No:2), by Hua Big Genomics is artificially synthesizing. After the synthetic primers were annealed, they were ligated with the linearized PX330 plasmid after digestion with BpiI, and the successfully constructed plasmid after ligation was named PX330-ROSA26 (the gene sequence of the plasmid is shown in SEQ ID No: 3). Simultaneously designed and synthesized ssODN-GFP reporter carrier by Nanjing GenScript Biotechnology Co., Ltd. (the gene sequence of the reporter carrier is shown in SEQ ID No: 4): a sequence homologous to the ROSA26 gene is inserted in the middle of ...
Embodiment 2
[0039] Example 2: Method for Improving the Efficiency of Site-directed Modification of Mouse Cell Genome
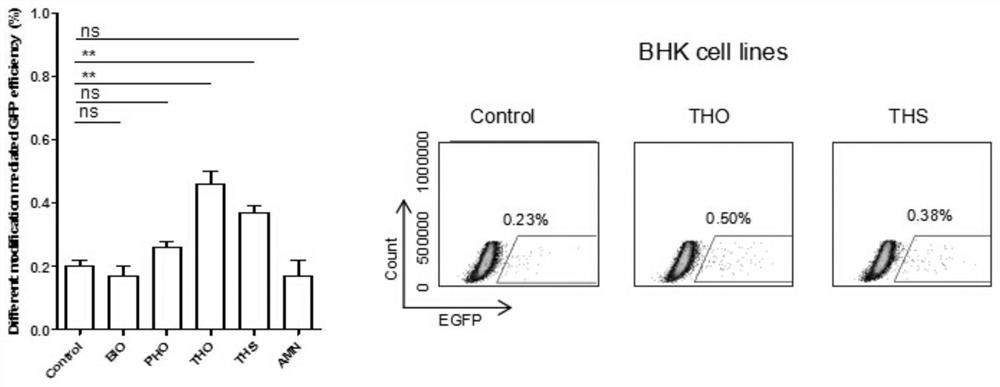
[0040] In this example, hamster kidney cells (BHK cells) were used as a tool cell line for experiments.
[0041] Wherein, the specific implementation steps of plasmid construction, cell recovery and culture, and tool cell line screening in the embodiment refer to steps 1-3 in the first embodiment.
[0042] 4. Results of co-transfection of PX330-ROSA26 plasmid and ssODN donor in hamster kidney cells
[0043] Transfection was started when the hamster kidney cells in the 24-well plate grew to 60%-80%. For the transfection process, refer to Thermofisher’s Instructions for LTX&Plus Reagent protocol, each well was transfected with 1 μg / well PX330-ROSA26 plasmid and 400ng ssODN. Among them, the transfected ssODNs were the unmodified blank group and the experimental group modified by BIO, PHO, THS, THO and AMN, respectively. After 4-6 hours, replace with fresh culture mediu...
Embodiment 3
[0045] Example 3: Method for Improving the Efficiency of Site-directed Modification of Porcine Cell Genome
[0046] In this example, pig-derived cells (PK-15 / PEF cell line) were used as a tool cell line for experiments.
[0047] Wherein, the specific implementation steps of plasmid construction, cell recovery and culture, and tool cell line screening in the embodiment refer to steps 1-3 in the first embodiment.
[0048] 4. Pig-derived cells co-transfected with PX330-ROSA26 plasmid and ssODN donor results
[0049] Transfection starts when the pig-derived cells (PK-15 / PEF cell line) in the 24-well plate grow to 60%-80%. The transfection process refers to Thermo fisher's Instructions for LTX&Plus Reagent protocol, each well was transfected with 1 μg / well PX330-ROSA26 plasmid and 400ng ssODN. Among them, the transfected ssODNs were the unmodified blank group and the experimental group modified by BIO, PHO, THS, THO and AMN, respectively. After 4-6 hours, replace with fresh c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com