Leveraging sequence-based fecal microbial community survey data to identify composite biomarker for colorectal cancer
A colorectal cancer and microbiological technology, applied in the field of markers for the diagnosis of CRC and CRA, and detection of transition from adenoma to cancer, can solve problems such as expensive, lack of comparability, and limited compliance rate of screening recommendations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
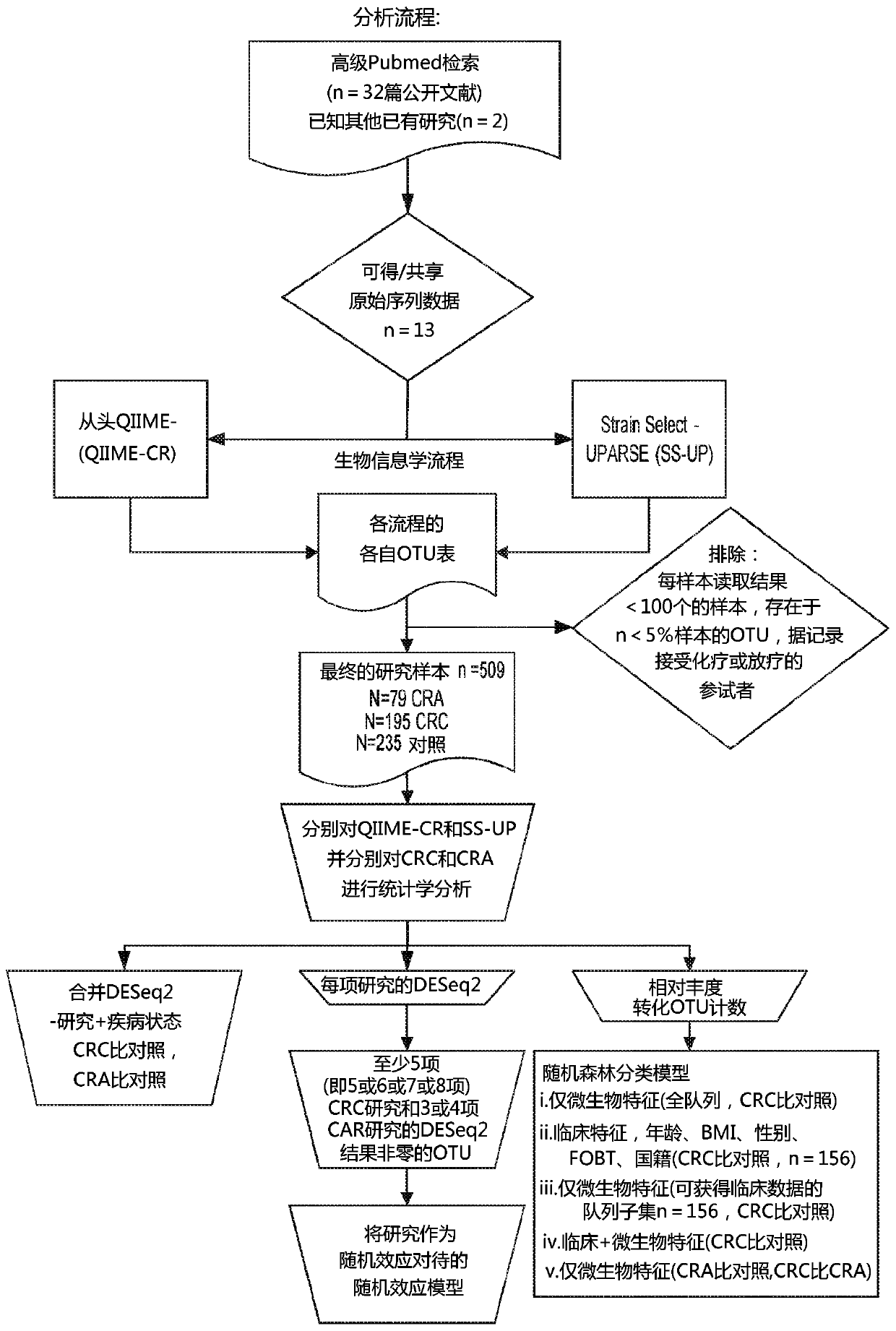
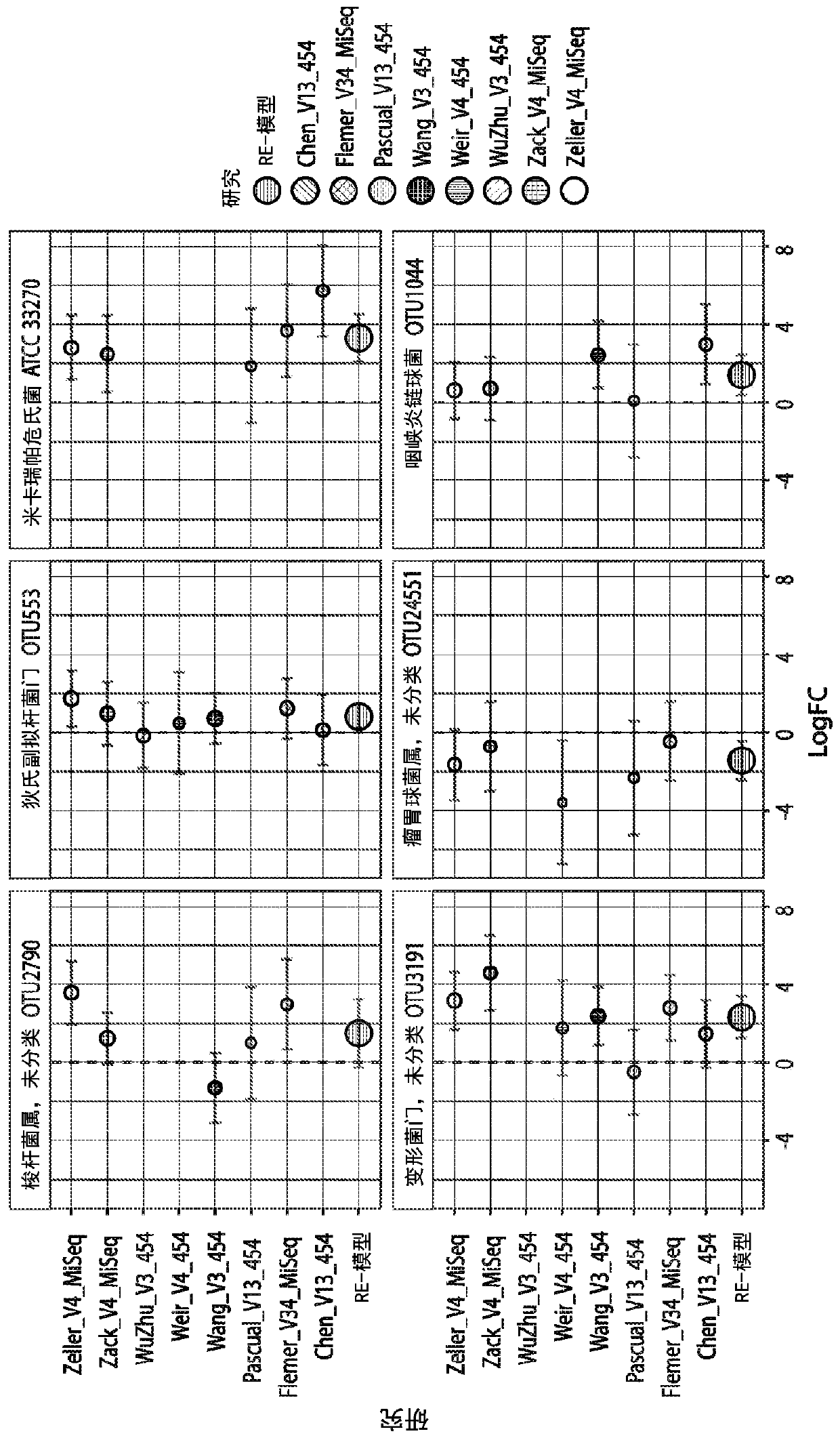
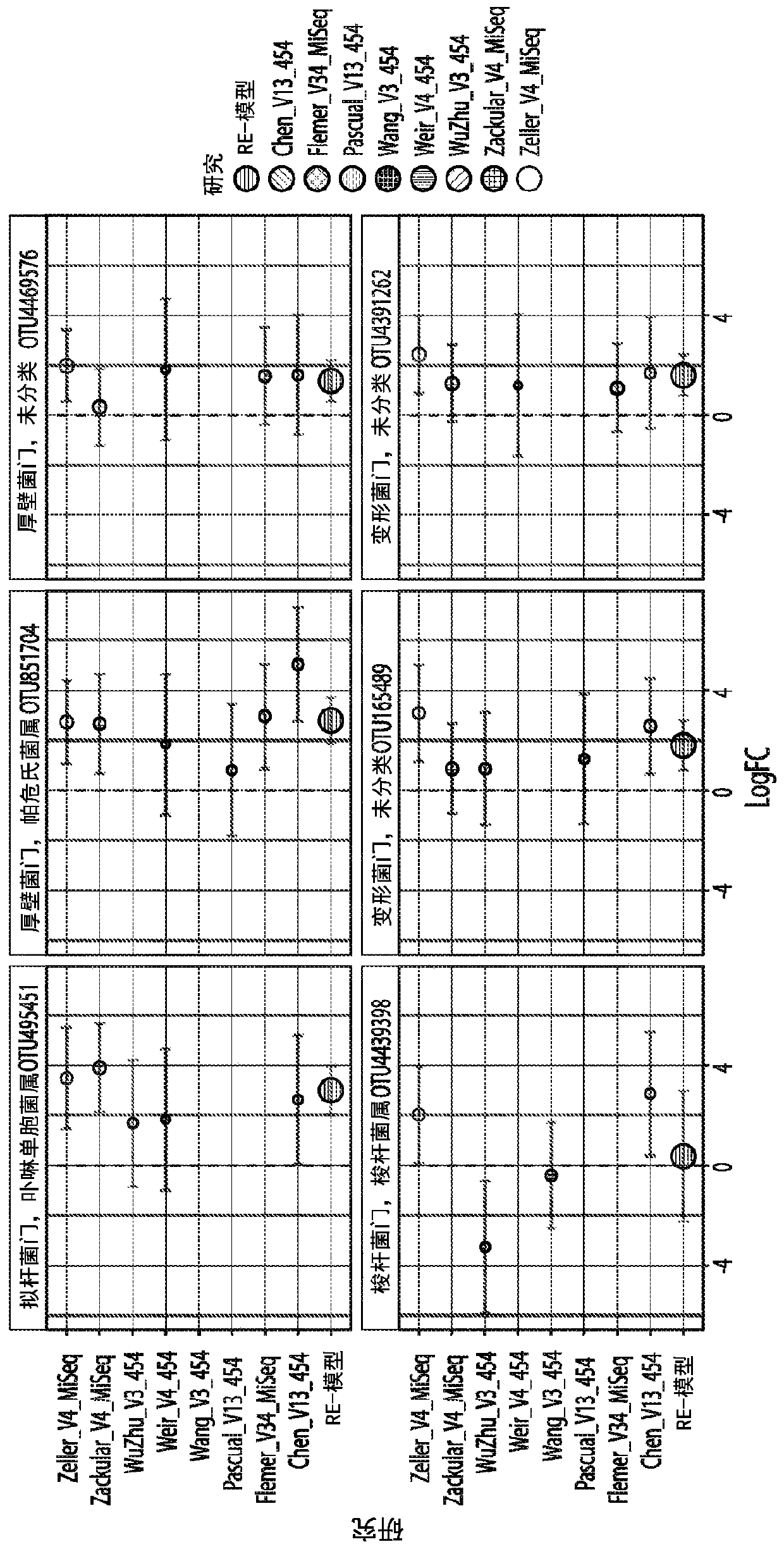
[0123] To determine whether universal microbial markers for CRC and CRA could be identified, we obtained raw 16S rRNA gene sequence data from multiple faecal microbiome studies published between 2006 and 2016. We analyzed the data using two bioinformatics pipelines, (1) de novo QIIME (QIIME-CR), a de novo (closed-reference ) OTU assignment method, and (2) Strain Select-UPARSE (SS-UP), a strain-specific method that in some cases uses more raw sequence data to give strain-level resolution. In addition, we compared our panel of microbial markers with the take-home guaiac-based fecal occult blood test (FOBT), a non-invasive but less precise test, where data were available. test. [23,24]
[0124] Research search, selection and adoption
[0125]We performed a systematic PubMed search for studies with colorectal cancer, colon cancer, or colorectal adenocarcinoma in the title, including human subjects, published between 2006 and 2016. The specific search terms that ended up using ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com