Gene methylation panel and kit for diagnosing and predicting curative effect and prognosis of colorectal cancer
A colorectal cancer and methylation technology, applied in the field of gene epigenetics, can solve the problems of colorectal cancer detection methods such as convenience in operation, accuracy of results and cost of detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] This example discloses a gene methylation panel for diagnosing and predicting the curative effect and prognosis of colorectal cancer. The gene methylation panel is composed of the first group of methylated genes, these methylated genes are MYO1G, BMPR1A , CD6, RBP5, Chr 13:10, LGAP5, ATXN1 and Chr 8:20. This gene methylation panel detects the gene methylation level changes of MYO1G, BMPR1A, CD6, RBP5, Chr 13:10, LGAP5, ATXN1 and Chr 8:20 in the plasma of colorectal cancer patients, and is used to diagnose the occurrence of colorectal cancer .
[0074] This embodiment also discloses the detection method of the above-mentioned gene methylation panel for diagnosing colorectal cancer, comprising the following steps:
[0075] In the first step, ctDNA is extracted from plasma;
[0076]In the second step, the extracted ctDNA is subjected to sulfite conversion, so that unmethylated cytosine in the ctDNA is deaminated and converted into uracil, while the methylated cytosine re...
Embodiment 2
[0092] The difference between this example and Example 1 is that the methylated genes in the gene methylation panel of this example are MYO1G, BMPR1A, CD6, Chr 13:10, LGAP5, ATXN1 and Chr 8:20.
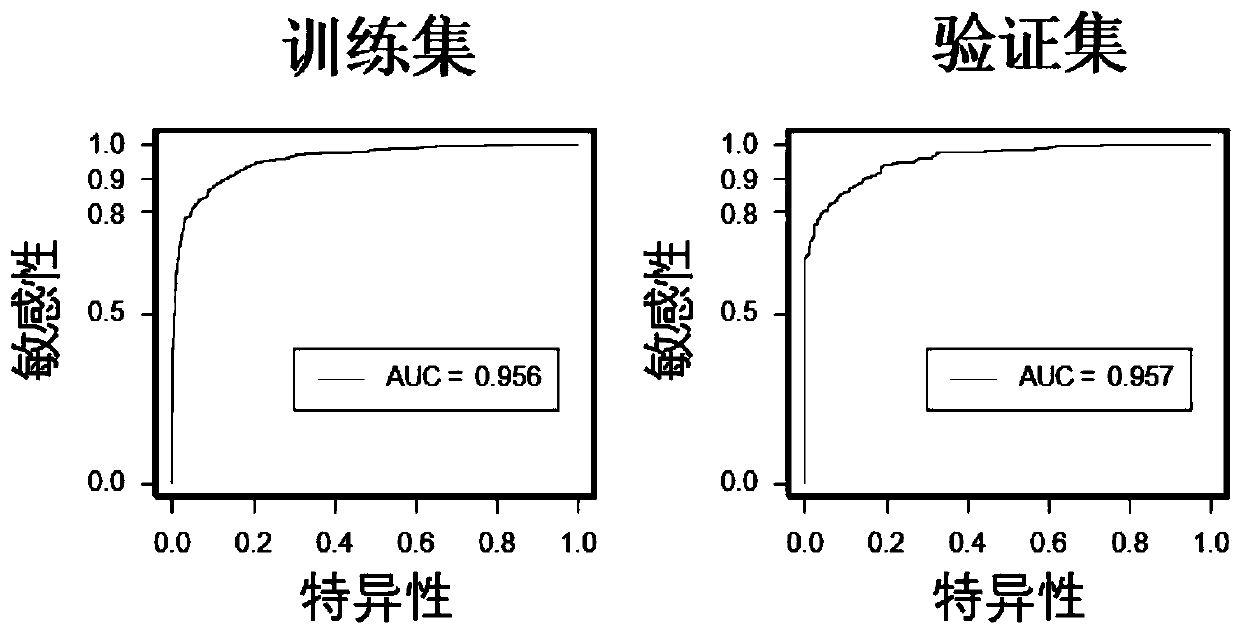
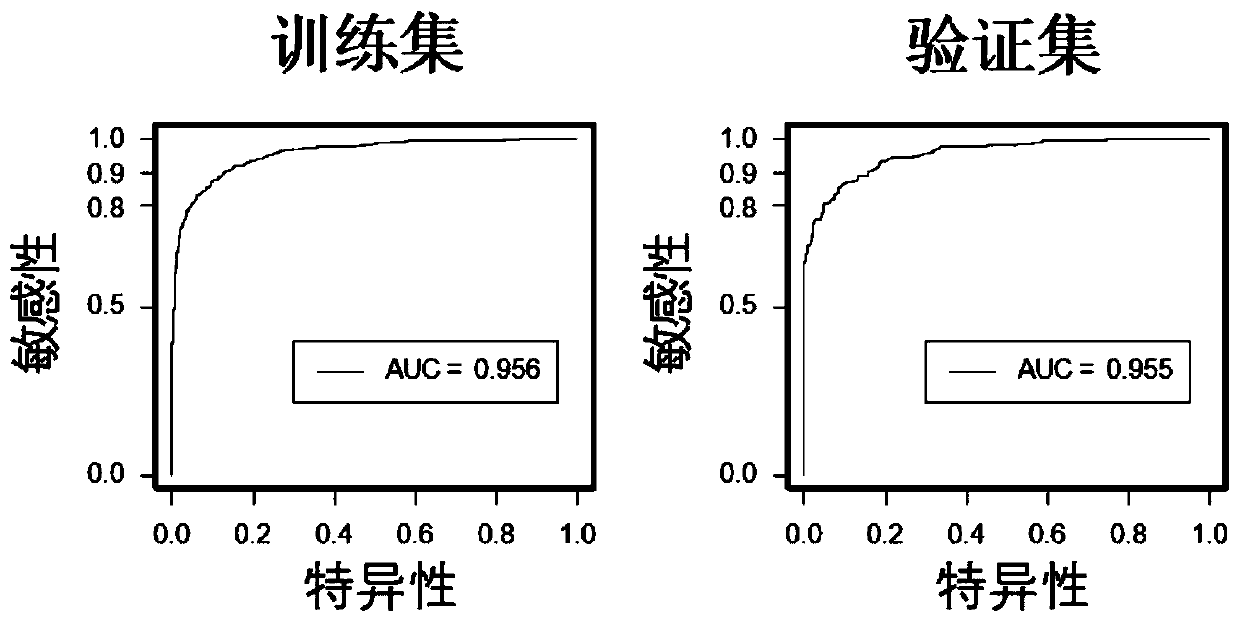
[0093] A diagnostic prediction model was constructed using a gene methylation panel (including 7 methylated genes: MYO1G, BMPR1A, CD6, Chr 13:10, LGAP5, ATXN1, and Chr 8:20) by logistic regression method. Using this model to diagnose colorectal cancer, the sensitivity in the training set is 87.69%, the specificity is 89.91%, and the overall correct rate is 88.94%. This model can very well distinguish colorectal cancer from normal controls in the training data set (AUC=0.956) and validation data set (AUC=0.955) (such as figure 2 , Table 3). It was confirmed that this gene methylation panel can be used to distinguish colorectal cancer patients from normal people.
[0094] Table 3: Sensitivity and specificity results of the gene methylation panel in the training set of Example 2
[0...
Embodiment 3
[0098] The difference between this example and Example 1 is that the methylated genes in the gene methylation panel of this example are MYO1G, BMPR1A, CD6, LGAP5, ATXN1 and Chr 8:20.
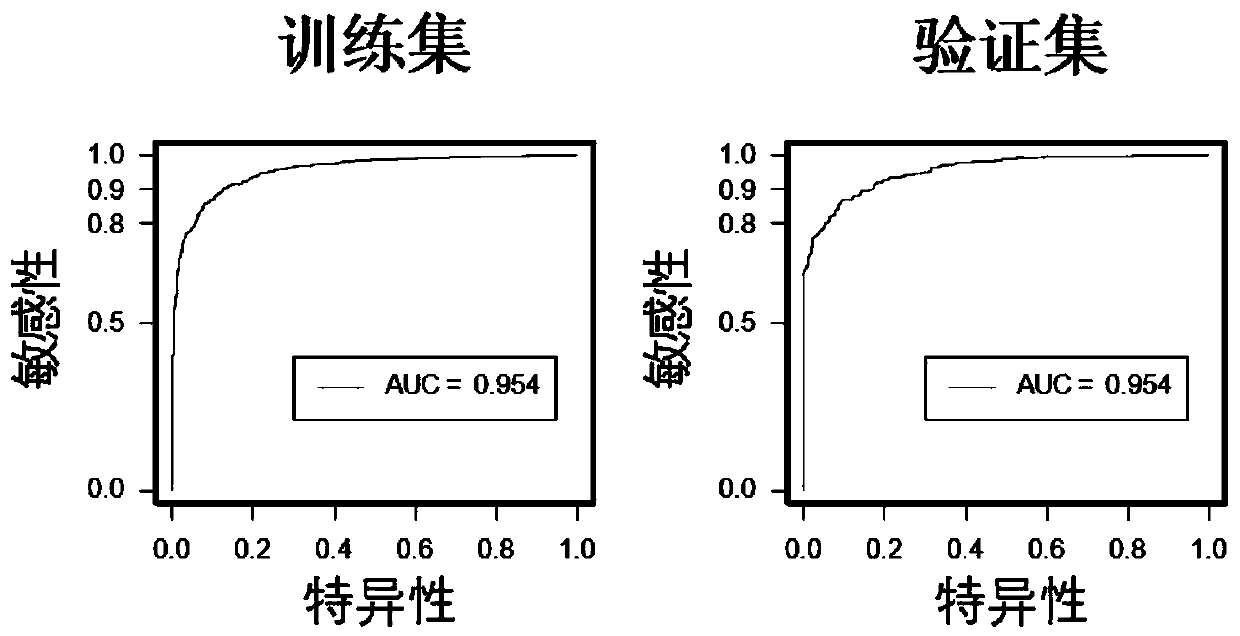
[0099] A diagnostic prediction model was constructed using a gene methylation panel (including 6 methylated genes: MYO1G, BMPR1A, CD6, LGAP5, ATXN1, and Chr 8:20) by logistic regression method. Using this model to diagnose colorectal cancer, the sensitivity in the training set is 85.61%, the specificity is 92.14%, and the overall correct rate is 89.27%. This model can very well distinguish colorectal cancer from normal controls in the training data set (AUC=0.954) and validation data set (AUC=0.954) (such as image 3 , Table 4). It was confirmed that this gene methylation panel can be used to distinguish colorectal cancer patients from normal people.
[0100] Table 4: Sensitivity and specificity results of the example three gene methylation panels in the training set
[0101]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com