Method for investigating molecules such as nucleic acids
A nucleic acid analysis and molecular technology, applied in the field of molecular research such as nucleic acid, can solve the problem of the limitation of the degree to which the speed at which droplets can move can be changed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0068] The method of the invention suitable for sequencing a DNA analyte according to a preferred third aspect of the invention is now illustrated by the following figures and the associated description.
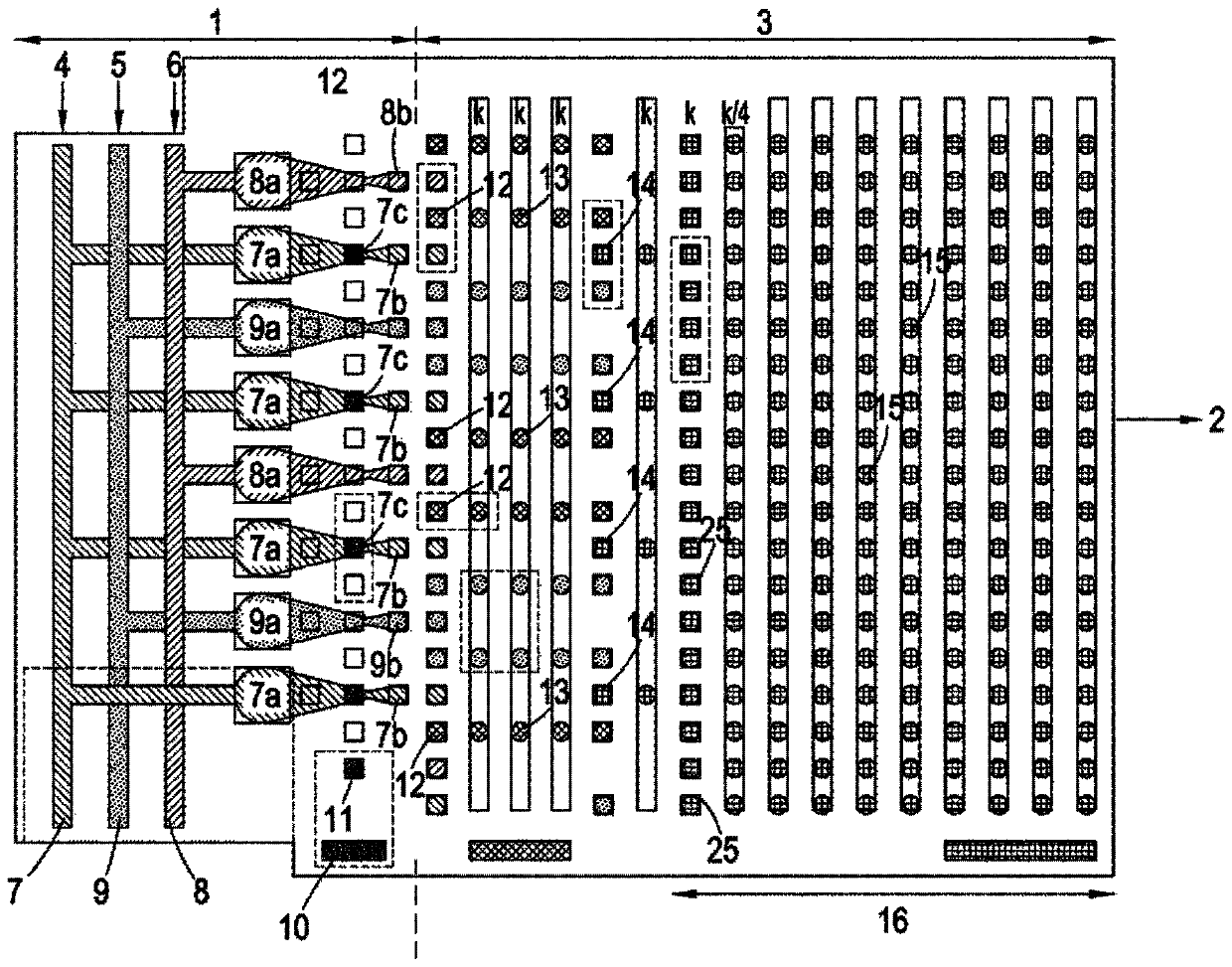
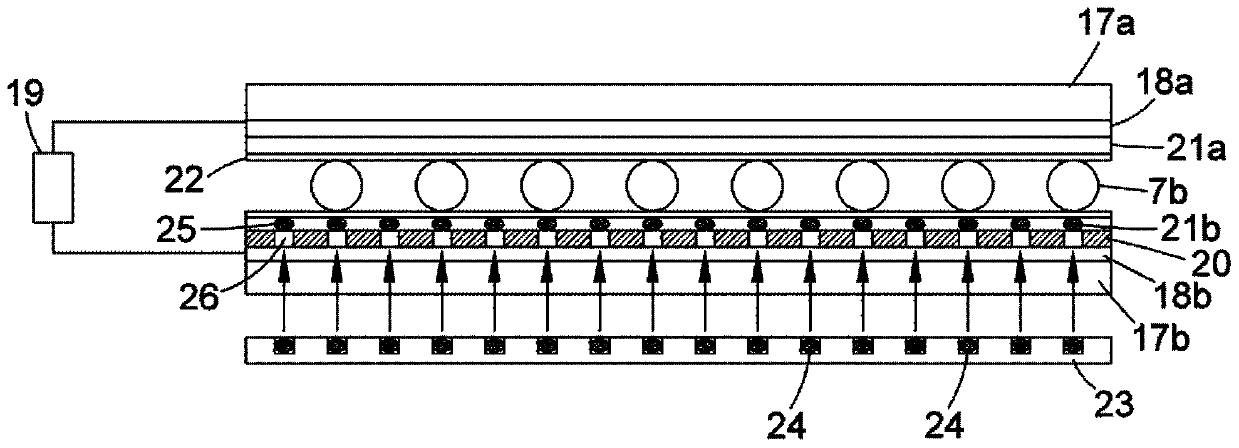
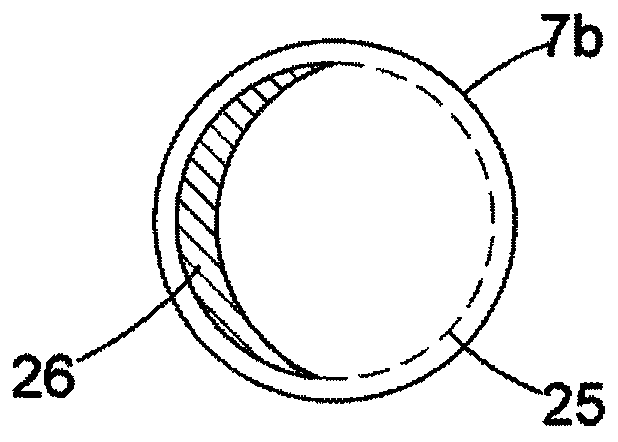
[0069] figure 1 shows a top view of a microfluidic substrate comprising a droplet preparation zone 1 and a droplet manipulation zone 3 integrated into a single substrate. 1 includes regions 7, 8 and 9 containing fluids attached to inlets 4, 5 and 6 which respectively introduce into the substrate a flow of pyrophosphate hydrolysis, a flow of inorganic pyrophosphatase, and various Detects the flux of chemicals and enzymes required to identify nucleoside triphosphates according to one of our earlier patent applications. These streams are then delivered to orifices 7a, 8a and 9a, respectively, from which various droplets 7b, 8b and 9b are produced (eg, using a droplet dispense head or by cutting out from larger intermediate droplets). 10 is a reservoir containing paramagnetic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com