Method and device for detecting human genome virus integration site
A technology for integrating sites and genomes, applied in genomics, proteomics, instruments, etc., can solve problems such as complex construction process, complex and cumbersome processing, and difficult processing, so as to achieve low requirements for computing resources, reduce the probability of mismatch, Fast running effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
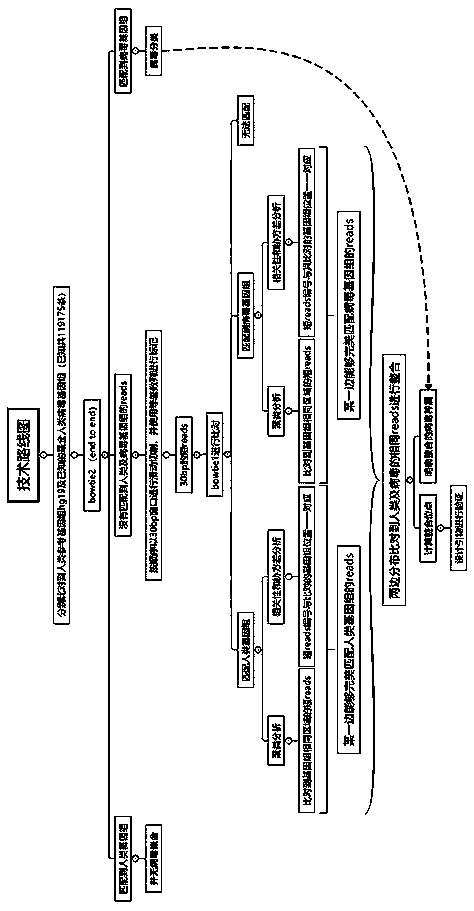
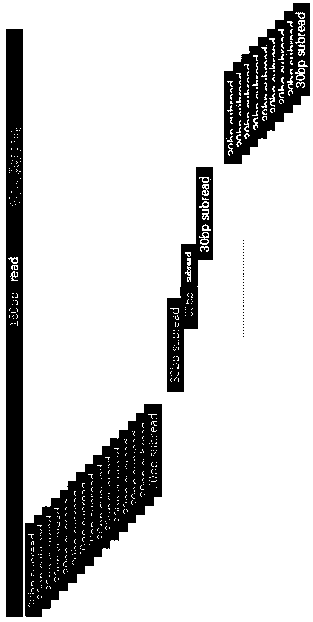
[0045] A method for detecting the integration site of human genome virus, according to the following steps, the technical route is as follows figure 1 Shown.
[0046] 1. Pre-treatment.
[0047] In order to ensure that the fastq sequence used for comparison meets the quality requirements, fastq data quality control filtering software is required, and fastp software is used to filter the fastq files obtained by sequencing result data.
[0048] 2. Genome comparison.
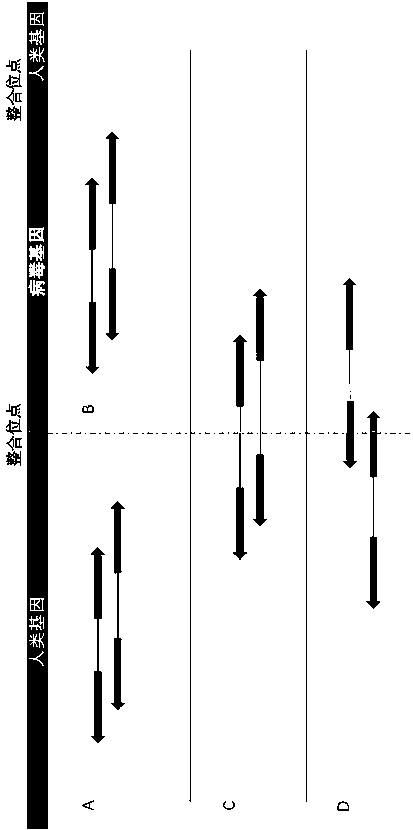
[0049] Such as figure 2 As shown, when the virus is integrated into the human genome, there are four situations in which reads obtained by sequencing:
[0050] A: Paired reads can be fully aligned to the human genome.
[0051] B: Paired reads can be fully aligned to the viral genome.
[0052] C: One pair of reads can be fully aligned to the human genome, and the other can be fully aligned to the viral genome.
[0053] D: One pair of reads can be completely aligned to the human genome or viral genome, while the other just falls ...
Embodiment 2
[0271] A device for detecting integration sites of human genome viruses includes a data acquisition module, an analysis module and an output module.
[0272] The data acquisition module is used to acquire data obtained from genetic testing.
[0273] The analysis module analyzes the human genome virus integration sites according to the method described in Example 1, and performs virus species analysis to obtain the inserted virus name.
[0274] The output module is used to output and display the results obtained by the analysis module.
Embodiment 3
[0276] According to the method of Example 1, the method of soft-clipping reads was used to analyze and process with HGT-ID software for comparison and verification.
[0277] 1. Method.
[0278] The same off-machine data for genetic testing was used, and analyzed according to the method in Example 1 and HGT-ID software respectively.
[0279] 2. Results.
[0280] (1) The method analysis result of Example 1.
[0281] Table 1. Output 1
[0282] ReadsID chr:start-end:chain Insertion site Virus type SRR1609136-17458832 chr8:128230627-128230679:1 chr8:128230627 Alphapapillomavirus 7 SRR1609136-25424758 chr8:128230627-128230680:1 chr8:128230627 Alphapapillomavirus 7 SRR1609136-26787992 chr8:128230627-128230678:1 chr8:128230627 Alphapapillomavirus 7 SRR1609136-30119552 chr8:128230627-128230675:1 chr8:128230627 Alphapapillomavirus 7 SRR1609136-74414072 chr8:128230627-128230674:1 chr8:128230627 Alphapapillomavirus 7 SRR1609137-32617125 chr8:128230627-128230678:1 chr8:128230627 Alphapapil...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com