Novel specific molecular targets for identifying salmonella and rapid detection method thereof
A technology of Salmonella and molecular targets, which is applied in the field of microbiological testing, can solve the problems of 13 specific new molecular targets of Salmonella that have not yet been discovered, such as the PCR detection method report, and achieve the effects of practicability, short detection time and low detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Mining of Salmonella Specific New Molecular Targets
[0035] Based on the GenBank database in NCBI and the whole genome DNA sequence of Salmonella self-tested by our team, comparative genomics analysis was carried out. The pan-genome analysis software Roary was used to cluster the functional genes with a similarity of 95% into the same gene cluster, and the comparison process was implemented in a localized script written in the Perl programming language. Convert the matrix file gene_presence_absence.csv generated after the pan-genome analysis into 0 / 1 format. Each column corresponds to the carrying status of the gene cluster of the strain. A value of 1 means that the gene is carried, and a value of 0 means that the gene is not carried. The unique gene fragments of Salmonella can be screened out by combinatorial sorting, and the gene sequences are shown in SEQ ID NO.1-13. The protein sequences of 13 specific genes of Salmonella strains were extracted, and comp...
Embodiment 2
[0036] Example 2 Establishment of a Rapid Detection Method for Salmonella Specific New Molecular Targets
[0037] This embodiment provides a method for detecting Salmonella: primers designed for any new specific molecular target of Salmonella obtained in Example 1 can form a rapid detection method, and include the following steps:
[0038] Design of primers: design specific amplification primers according to the above-mentioned 13 new specific molecular target base sequences SEQ ID NO.1-13, and the primer sequences are as follows:
[0039] Table 1 Salmonella specific PCR detection primer sequence
[0040]
[0041]
[0042]
[0043] DNA template preparation: Bacteria were cultured in nutrient broth, and bacterial genomic DNA was extracted using a commercial bacterial genomic DNA extraction kit as a template to be tested.
[0044] PCR detection system and amplification procedure:
[0045] The PCR detection system is 25 μL, including: 2×PCR Mix 12.5 μL, template DNA 1 ...
Embodiment 3
[0055] The detection of embodiment 3 Salmonella suspected bacterial strains
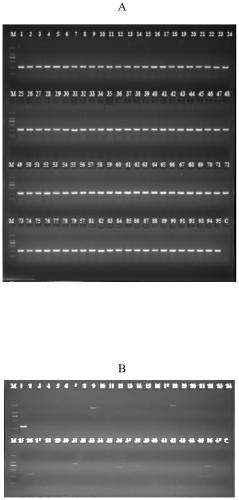
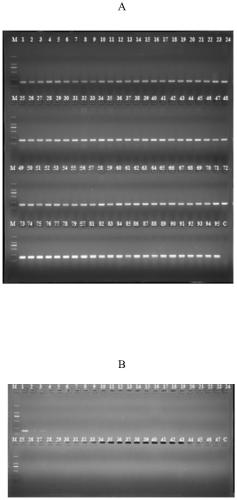
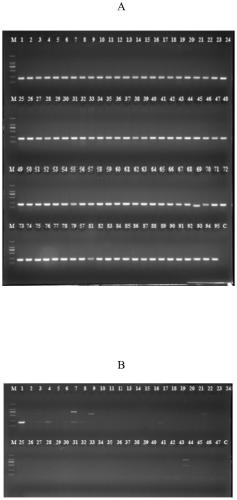
[0056] The Salmonella PCR detection method in Example 2 is used to detect 391 suspected strains of Salmonella isolated from food samples. The food samples are collected from supermarkets and bazaars in Guangdong Province. For sample processing and the isolation of suspected bacterial strains, refer to the national standard method. Using the specific molecular target, primer and detection method of the present invention, a total of 391 strains of Salmonella were detected, which was consistent with the biochemical reaction test and 16s rDNA identification results. This example illustrates that the specific target of Salmonella of the present invention and the detection primers are highly reliable.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com