Cancer staging prediction system based on genome analysis
A genome analysis and prediction system technology, applied in the field of cancer staging prediction system, can solve problems such as network complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
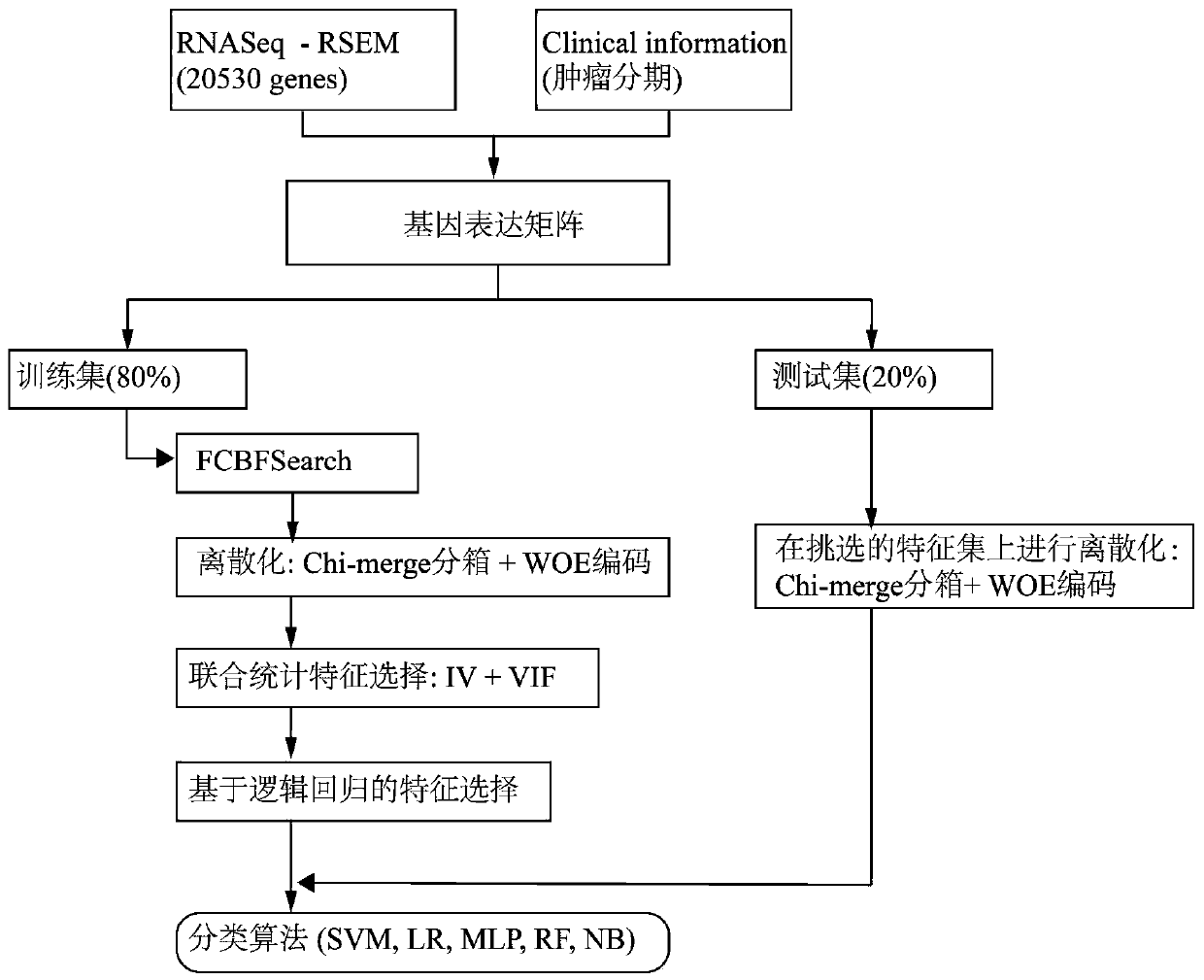
[0083] A cancer staging prediction system based on genomic analysis, including a raw data acquisition unit, a combined feature preprocessing unit, a joint gene selection unit, a classification model creation unit, and a prediction unit connected in sequence;
[0084] The raw data acquisition unit is used to: obtain the RNAseq expression data and clinical information of the cancer subtype samples corresponding to the Cancer Genome Atlas TCGA project, obtain the RSEM value of the gene expression in it, and samples with phase I and phase II annotations are considered early cancers, and the rest Samples with stage III and IV annotations are advanced cancers;
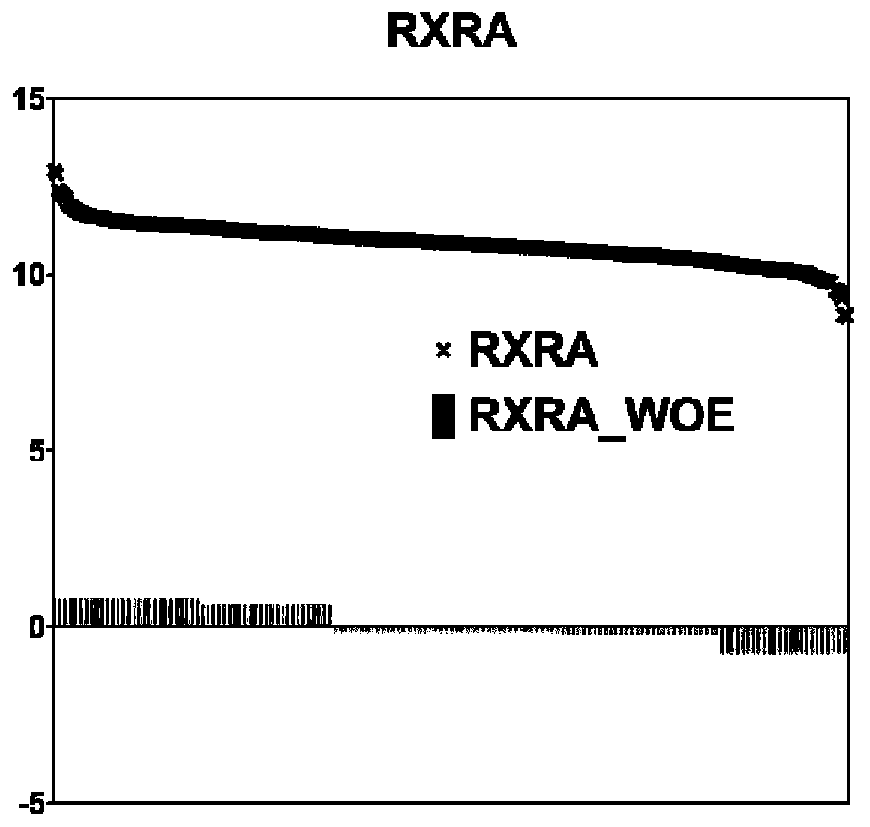
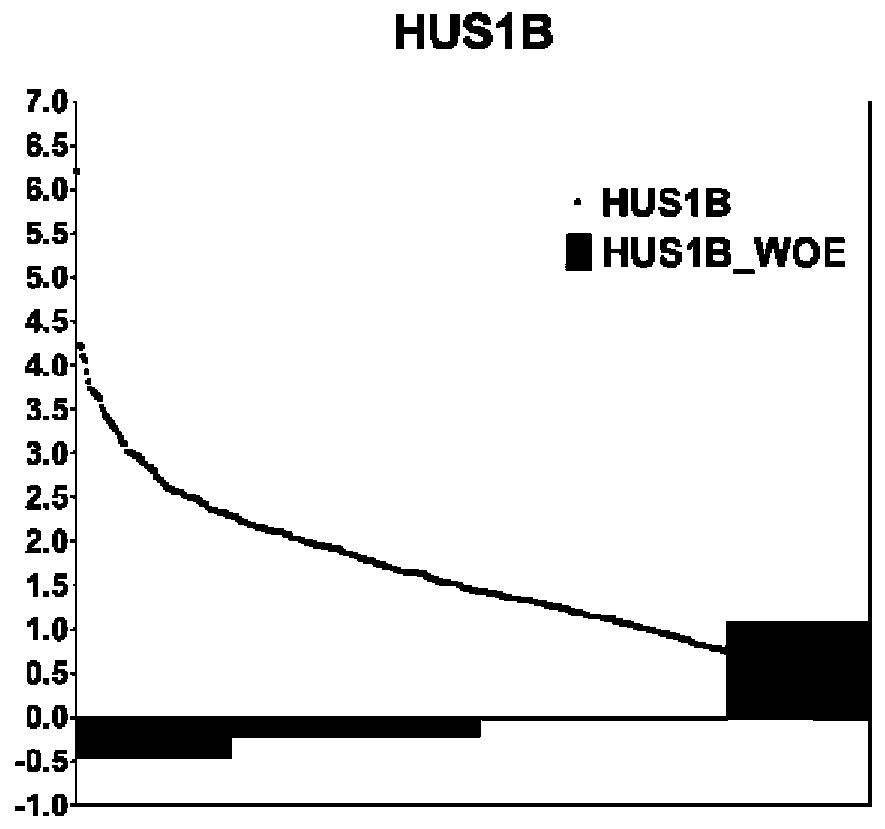
[0085] The combined feature preprocessing unit is used to discretize genetic features, that is, RNAseq expression data, through ChiMerge binning and WOE encoding, and improve the stability of the data and the robustness of the classification model through ChiMerge binning and WOE encoding.
[0086] ChiMerge binning: Through ...
Embodiment 2
[0105] According to a cancer stage prediction system based on genomic analysis described in Example 1, using log2 to convert the RSEM value, and standardizing the RSEM value after log2 conversion refers to:
[0106] Use log2 to transform the RSEM value by formula (I):
[0107] x=log 2 (RSEM+1) (I)
[0108] Standardize the RSEM value after log2 transformation by formula (II) to get z:
[0109]
[0110] In formula (II), x is the logarithmized value of RSEM value, is the mean of x and s is the standard deviation.
[0111] FCBF search is performed on the original training data, which refers to RSEM values, including:
[0112] (1) Use random sampling to select 80% of the original training data as the training data set. In the ten times of ten-fold cross-validation experiments, the training data is randomly divided into ten folds each time, and the FCBF search is performed on the training data set. , each time FCBF searches for ten-fold cross-sampling, and obtains 10 sub-fe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com