A universal high-throughput sequencing linker and its application
A sequencing linker and high-throughput technology, which can be used in the detection/testing of microorganisms, library creation, biochemical equipment and methods, etc., and can solve the problems that different sequencing platforms cannot be realized
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0150] Example 1 Design and preparation of high-throughput sequencing adapters
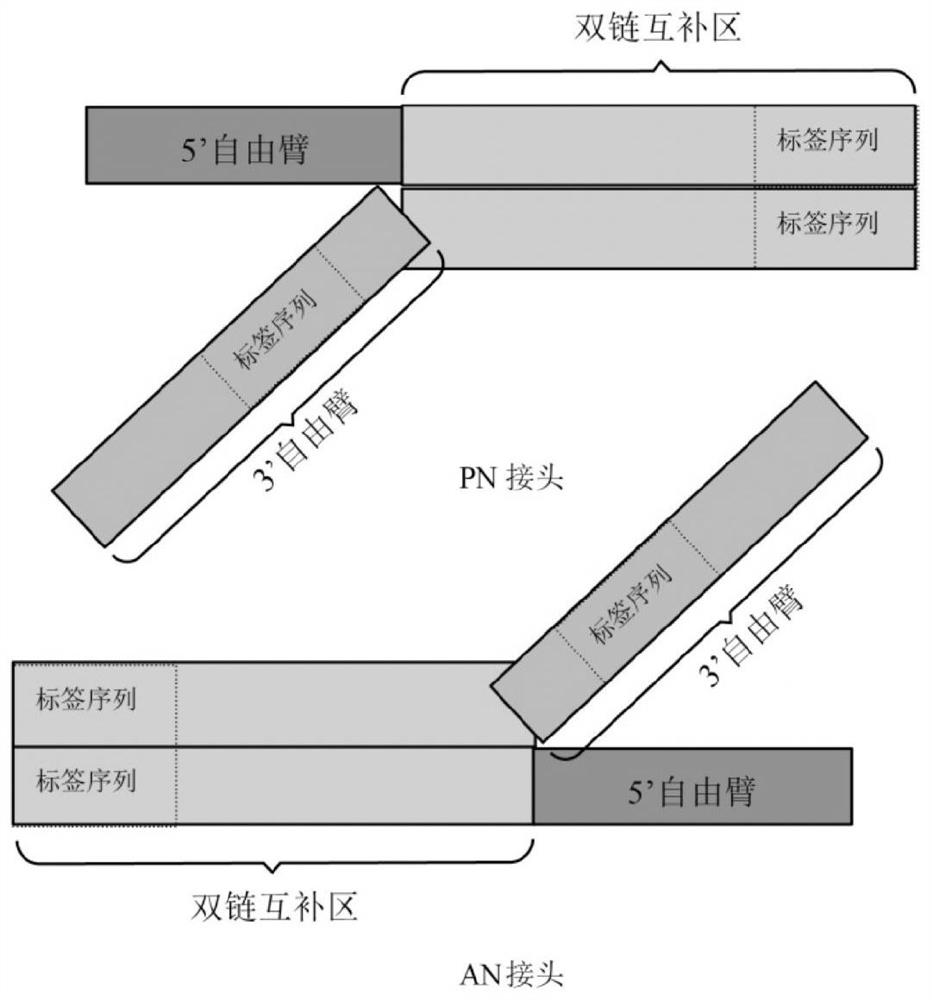
[0151] according to figure 1 As shown in the structural composition, two sets of universal high-throughput sequencing adapters AN1 / PN1 and AN2 / PN2 were designed. Among them, AN1 / PN1 is a set of universal high-throughput sequencing adapters with short sequences, and AN2 / PN2 is another set of universal high-throughput sequencing adapters with longer sequences.
[0152] The specific preparation method is as follows:
[0153] The sequencing adapters shown in the following sequences 1-4 were prepared, the sequences 1 and 2 were annealed to form the AN1 adapter, and the sequences 3 and 4 were annealed to form the AN2 adapter.
[0154] Sequence 1:
[0155] 5'-XXXXXXTAGCTGAGTCGGAGACACGCAGGATCGGAAGAGCACGTCTGAACTCCAGTCACXXXXXXATCTCGTA*T*G-3'; (3' free arm chain)
[0156] Sequence 2:
[0157] 5'-ACCGAGATTCTACACTCTTTCCCTACACGACGCTCTTCCGATCCTGCGTGTCTCCGACTCAGCTAXXXXXX-3'; (5' free arm chain)
[0158] Seq...
Embodiment 2
[0172] Example 2 General high-throughput sequencing library preparation of aorta-related genes
[0173] The universal high-throughput sequencing adapters AN1 / PN1 and AN2 / PN2 prepared in Example 1 were used for experiments.
[0174] Wherein, the number of universal high-throughput sequencing adapters corresponds to the number of samples to be tested. For example, if the number of samples to be tested is 10, 10 sets of universal high-throughput sequencing adapter sets are correspondingly prepared, and each set of universal high-throughput sequencing adapter sets includes PN1 adapters and AN1 adapters. The nucleotide sequences of the tag sequences in the same group of PN1 adapters and AN1 adapters are the same, and the base sequences of the tag sequences in different adapter groups are different.
[0175] Ligate the target fragment to be tested with the above sequencing adapter set to obtain a ligation product; wherein, the gene fragments from the same sample source are connecte...
Embodiment 3
[0202] Example 3 Sequencing analysis based on Ion Torrent platform and Illumina platform
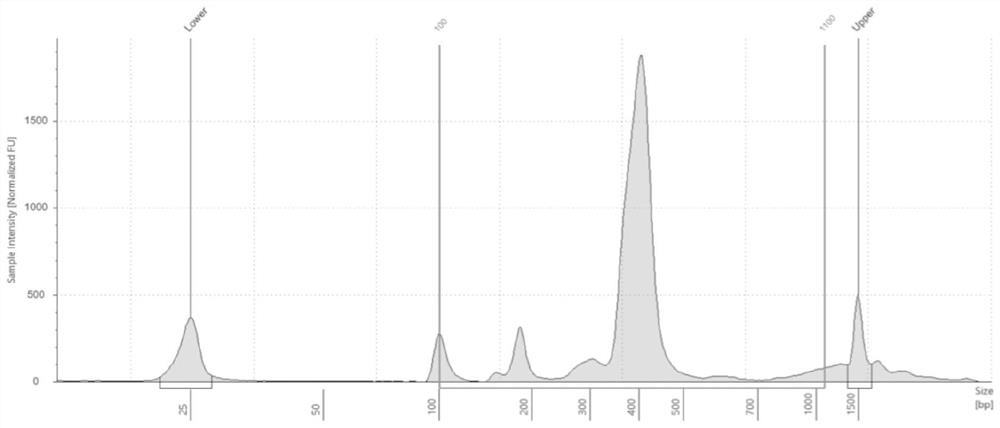
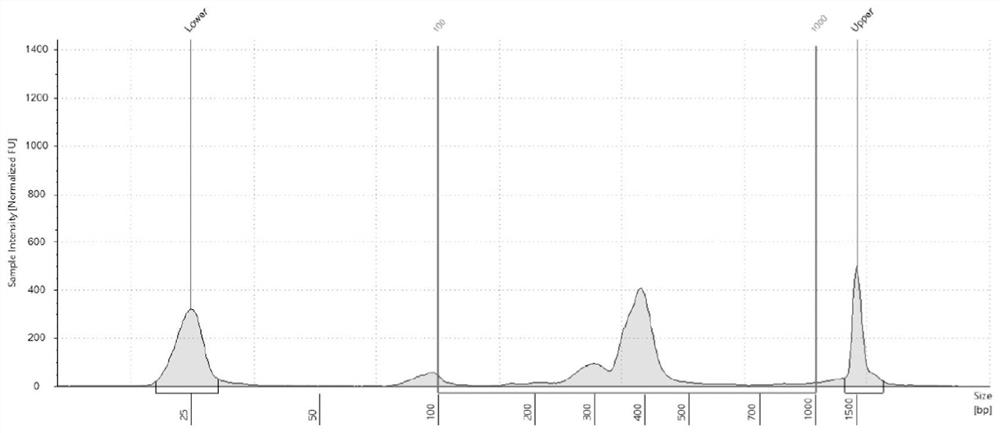
[0203] In this example, the Ion Torrent platform and the Illumina platform were used to perform sequencing verification on the above-mentioned high-throughput library, as follows:
[0204] 1. Ion Torrent platform Ion GeneStudio TM Sequencing on the S5 Plus sequencer, the specific implementation steps are as follows:
[0205] Dilute the library after the above purification and quality inspection, using Ion 520 TM &Ion 530 TM Kit–OT, according to the kit protocol, after template preparation on the IonTouch 2 instrument, Ion GeneStudio TM Sequencing and data analysis were performed on the S5 Plus gene sequencer.
[0206]2. Sequencing on the Illumina platform Miseq DX sequencer, the specific implementation steps are as follows:
[0207] After the above-mentioned purification and quality inspection, the library was diluted, and the Miseq DX Reagent Kit v3 was used to perform sequencin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com