Single-cell sequencing data dimension reduction method fusing gene ontology and neural network
A single-cell sequencing and gene ontology technology, applied in character and pattern recognition, instruments, biostatistics, etc., can solve the problems of mediocre effects, insufficient use of biological information knowledge, and weak interpretation, so as to achieve accelerated training and fast Effective dimensionality reduction and enhanced dimensionality reduction effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0029] The present invention will be further described below in conjunction with the accompanying drawings and embodiments, and the present invention includes but not limited to the following embodiments.
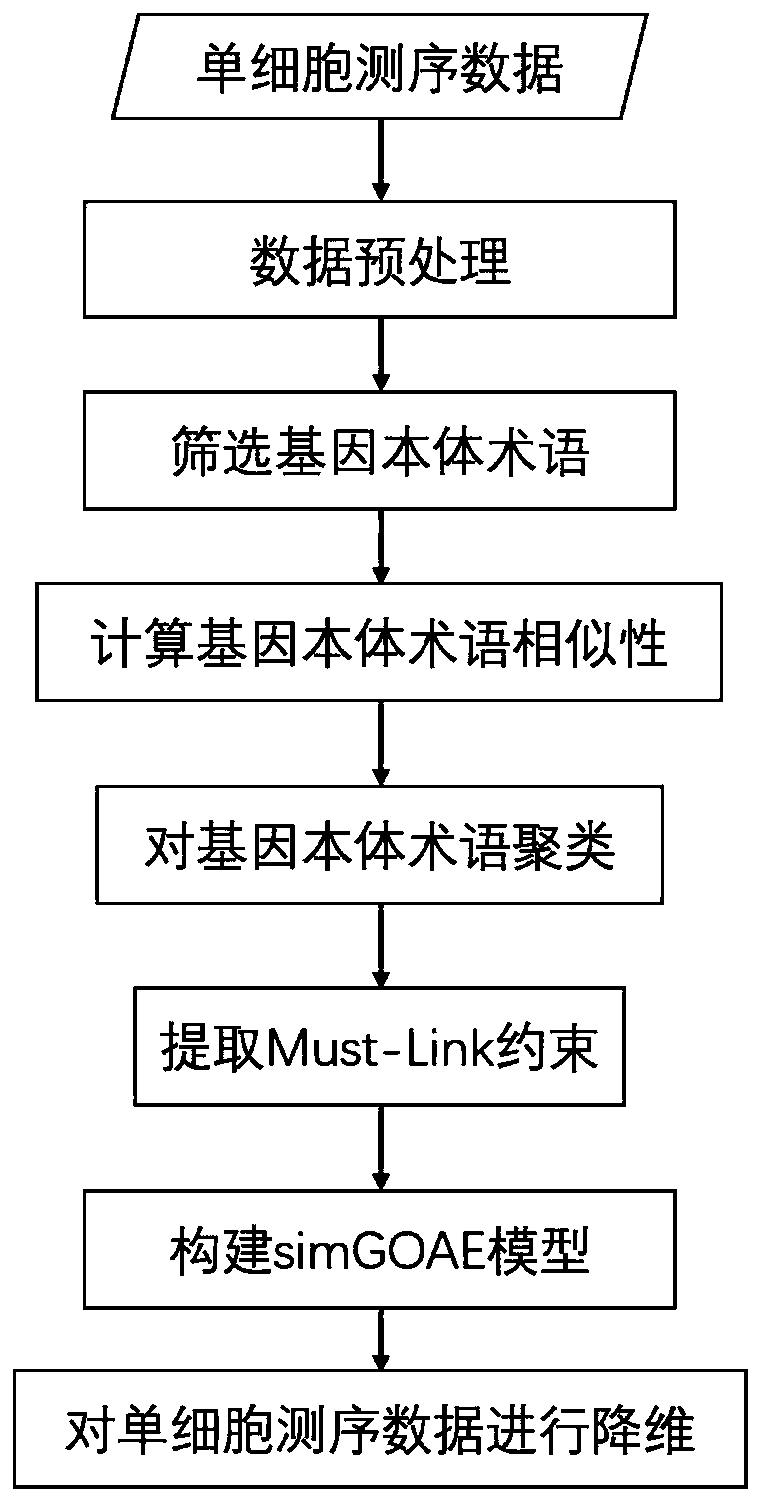
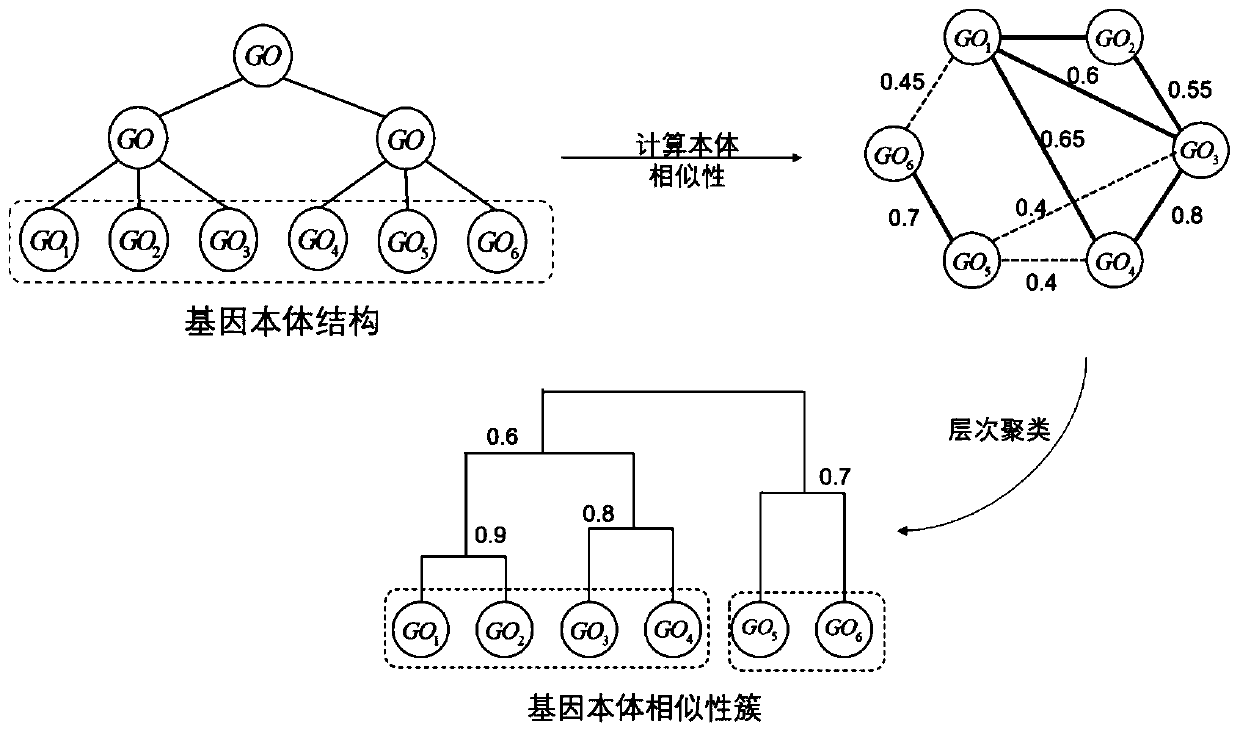
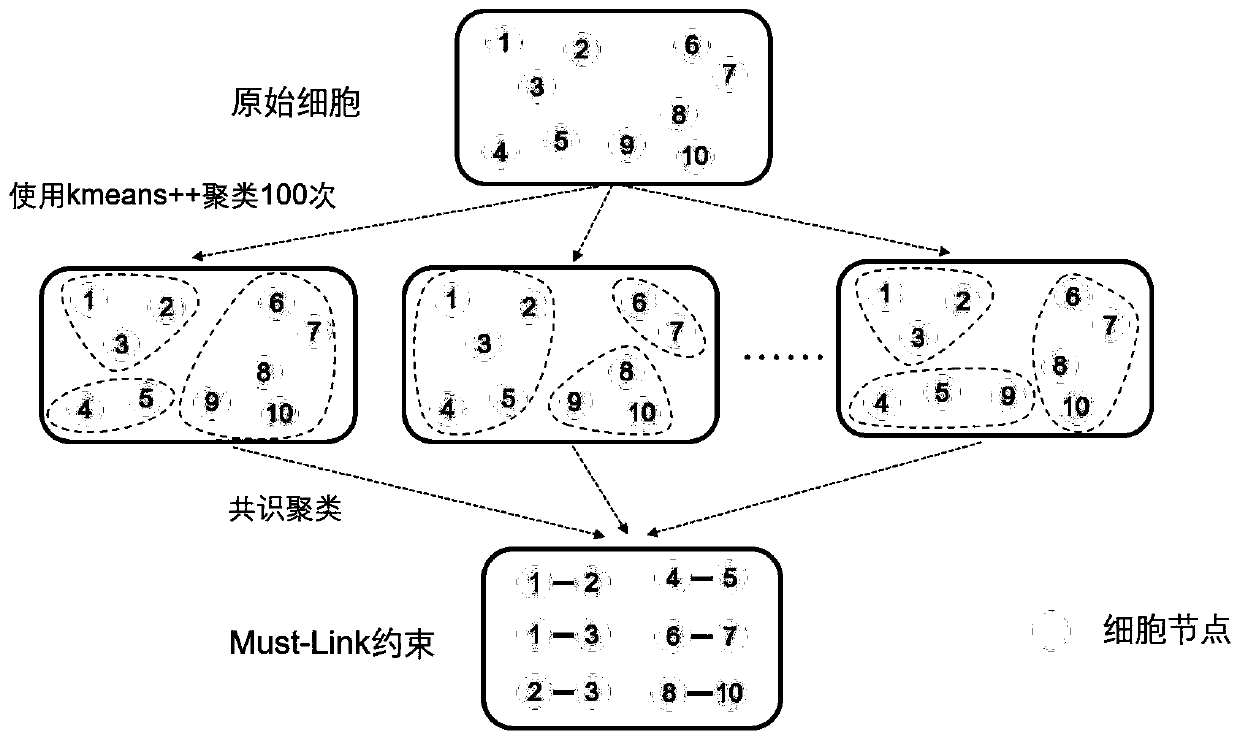
[0030] Single-cell sequencing data can be viewed as a matrix in which the horizontal and vertical coordinates are cells and genes, respectively. The numbers in the matrix represent the amount of gene expression in a certain cell, and are generally represented by real numbers. like figure 1 As shown, the present invention provides a single-cell sequencing data dimensionality reduction method that integrates gene ontology and neural network, and its basic implementation process is as follows:
[0031] 1. Data preprocessing
[0032] Generally, the original single-cell sequencing data are natural numbers, which are preprocessed.
[0033] (1) Delete the gene whose gene expression is less than 3 cells in the single-cell sequencing data (the gene expression value is 0, 1, 2... whe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com