A high-fidelity polymerase with gap DNA preference and its application
A high-fidelity polymerase and preference technology, applied in the field of genetic engineering, can solve the problems of no substrate preference and lack of application value of polymerase, and achieve the effect of avoiding degradation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Embodiment 1: Construction of Deinococcus radioduran KlenDr protein expression strain
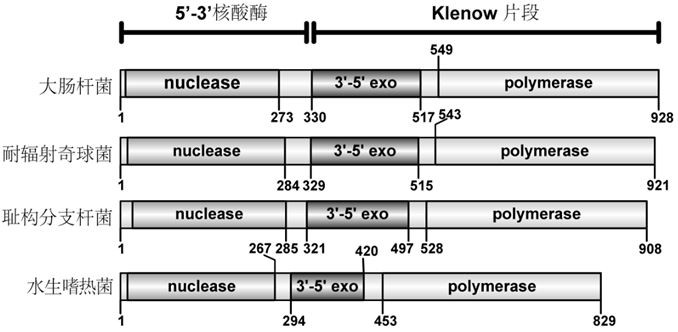
[0026] According to the annotated Deinococcus radiodurans R1 genome (ASM163882v1) resequenced in 2016, DNA polymerase I (Protein Sequence ID: ANC71194.1) contains 921 amino acids, which is 35 amino acids less at the N-terminus than the old version (ASM856v1). The KlenDr involved in the present invention is derived from amino acids 289-921 of the new DNA polymerase I sequence. figure 1 is a schematic diagram of the domains of the DNA polymerase I family, in which the domain of Deinococcus radiodurans is located in the second item.
[0027] (1) The genome of Deinococcus radiodurans R1 was extracted using the Bacterial Genomic DNA Extraction Kit (DP302-02) of Tiangen Biotechnology, and its concentration and purity were determined with NanoDrop 1000 (Thermo Company).
[0028] (2) Design a pair of primers that can be used for homologous recombination according to amino acids 289-921 of t...
Embodiment 2
[0032] Embodiment 2: Induced expression of Deinococcus radiodurans KlenDr protein
[0033] (1) Transform the successfully constructed Pet28a-KlenDr expression vector into Escherichia coli BL21(DE3) expression strain (Quan Shi Jin Company), and use solid LB medium containing 40 μg / mL Kanamycin and 34 μg / mL chloramphenicol antibiotics to screen for successful transformation cloning strains.
[0034] (2) Pick a successfully transformed single colony into 5ml liquid medium, shake and culture overnight at 37°C, then transfer to 500ml medium, culture until OD600 is 0.6-0.8, put it on ice for 10min, add 100ul 1M IPTG was cultured at 30°C for 5h to induce the expression of the target protein.
[0035] (3) After induction, collect the cells by centrifugation at 8000rpm for 8 minutes, resuspend and wash the cells with 1x PBS, and finally centrifuge at 8000rpm for 5 minutes, discard the supernatant, and store the cells at -80°C.
Embodiment 3
[0036] Embodiment 3: the purification of Deinococcus radiodurans KlenDr protein
[0037] (1) Cell lysis: Add 15 mL of lysis buffer (300 mM NaCl, 20 mM Tris HCl pH 8.0, 5% Glycerol, 3 mM β-Me, 10 mM imidazole) to resuspend cells according to 1 g of cells (wet weight). Use an ultrasonic cell breaker (Ningbo Xinzhi, JY92-IIN) to break the cells in an ice-water bath until the suspension is transparent. The parameters are alternating rod Φ6, power 60%, ultrasound 2.5s, gap 9.9 s, time 60-90 min . The broken cell suspension was centrifuged at 15,000 rpm for 35 minutes to remove cell debris, and the supernatant was retained and filtered with a 0.22 or 0.45 μM filter membrane.
[0038](2) Nickel column affinity purification: Nickel column (HisTrap HP 1ml) was purchased from GE HealthCare. First use Ni-bufferA (300 mM NaCl, 20 mM Tris HCl pH 8.0, 5% Glycerol, 3 mM β-Me) to equilibrate the nickel column, use a flow rate of 1ml / min to fully combine the cell lysate with the nickel colum...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com