Specific primer pair, probe and detection kit for detecting carp herpesvirus type Ⅱ
A detection kit and primer pair technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of limiting the use of LAMP method, false positive results, etc., to avoid false positive results The effect of appearance, high accuracy and low energy consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
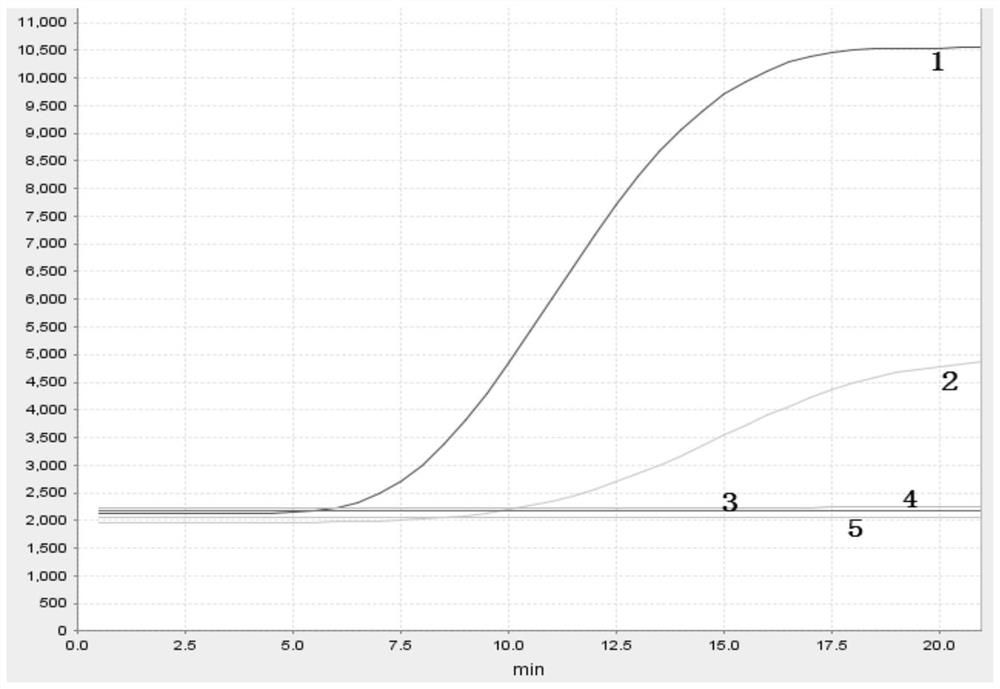
[0065] 1. Obtaining the positive plasmid of CyHV-2: the common CyHV-2 ORF71 gene listed in the NCBI search literature, using Vector NTI software to compare and find out its conserved region, select the ORF71 gene part region as the target amplification segment, It was constructed into the vector puc57 to prepare a CyHV-2 positive plasmid. The plasmid was synthesized by Sangon Bioengineering (Shanghai) Co., Ltd. The partial sequence of the selected ORF71 gene is as follows:
[0066] GTCAAGTGCGCCTCTTTCAACCTACCCTTTAGCGTCAGGTCCATAGAGGATCCAGAGTACAGCGAGTGTCTGGATATGGTTCAGCACAACGTTAGCACCGTACGTTTCCAAGAGATTATGCAGTCTCGGGTGAGGACTTGCGAAGAGTTTGATTTCTACACGCCTCGCATCATGCATCAGGACAACGCGGTCAGACAACTCAACGAGTCTTGTATGAAAAAGACTGTGGGCGCCGAACGGATCTTCAAGCCCAAGATCAATCACAATAACGTGCAGAACCCGGACGAGCGTAGAAAGTTTGCAGCCGTGGTCCGTCAACGGTTCAAGCACATTGACTTCTTTCAAGGCGTCCGAATCAAGGTCGGATCTCTGGTGTGCGTACTAAAATATCAAACTCAAGTGTTTGAAGGCTGTCTGGGAATAGTGGAATCAGTACAACCCGTCATGGTACGCCTTTTTTTGTTTGTTTGTTTGTTTGATGAGACGATGGTGACTATGGTTAATGT...
Embodiment 2
[0093] Select the following primers and probe sequences:
[0094] The upstream primer sequence is: 5'-CGCCTCTTTCAACCTACCCTTTAGCGTCAG-3' (SEQ ID NO: 3);
[0095] The downstream primer sequence is: 5'-GTCCGGGTTCTGCACGTTATTGTGATTGAT-3' (SEQ ID NO: 4);
[0096] The probe sequence is: 5'-TGTTTGTTTGTTTGTTTGATGAGACGATGG(BHQ1-dT)G(THF)C(FAM-dT)ATGGTTAATGTGTTGT(C3-SPACER)-3'(SEQ ID NO: 9)
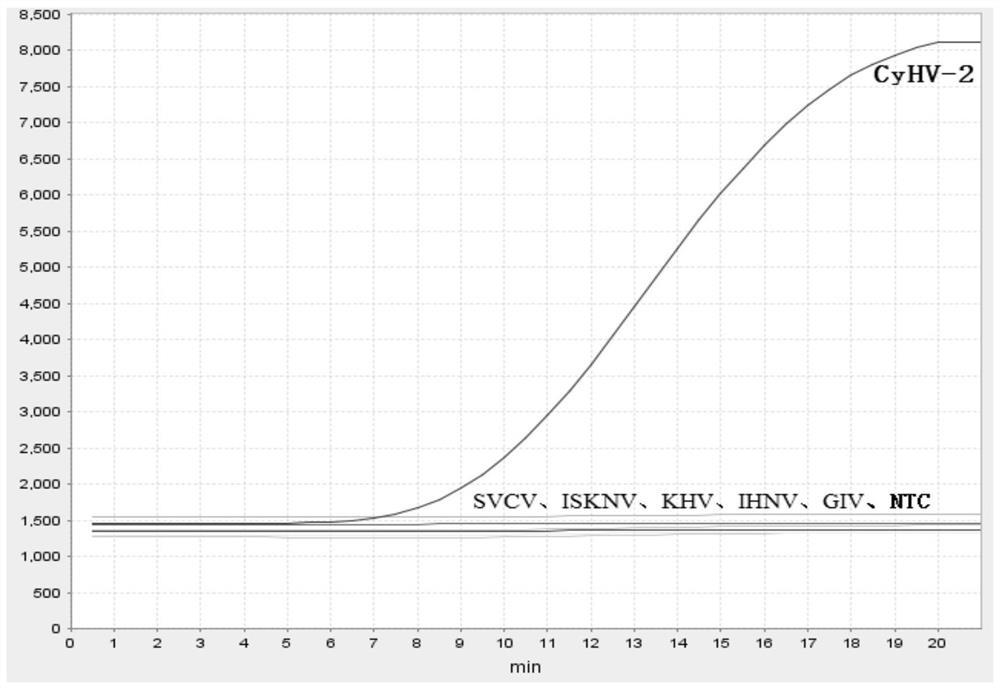
[0097] By synthesizing a plasmid containing the CyHV-2ORF71 gene sequence as the detection target, the method version amplification test of recombinase polymerase amplification (combined with endonuclease IV) was carried out, and the 50 μl amplification reaction system was constructed as follows:
[0098] 60mM tris-acetate buffer pH8.0
[0099] 100mM potassium acetate
[0100] 14mM magnesium acetate
[0101] 3mM Dithiothreitol
[0102] 5% polyethylene glycol (20000)
[0103] 2mM ATP
[0104] 20mM creatine phosphate
[0105] 100ng / μl creatine kinase
[0106] 600ng / μl E. coli SSB protein
[...
Embodiment 3
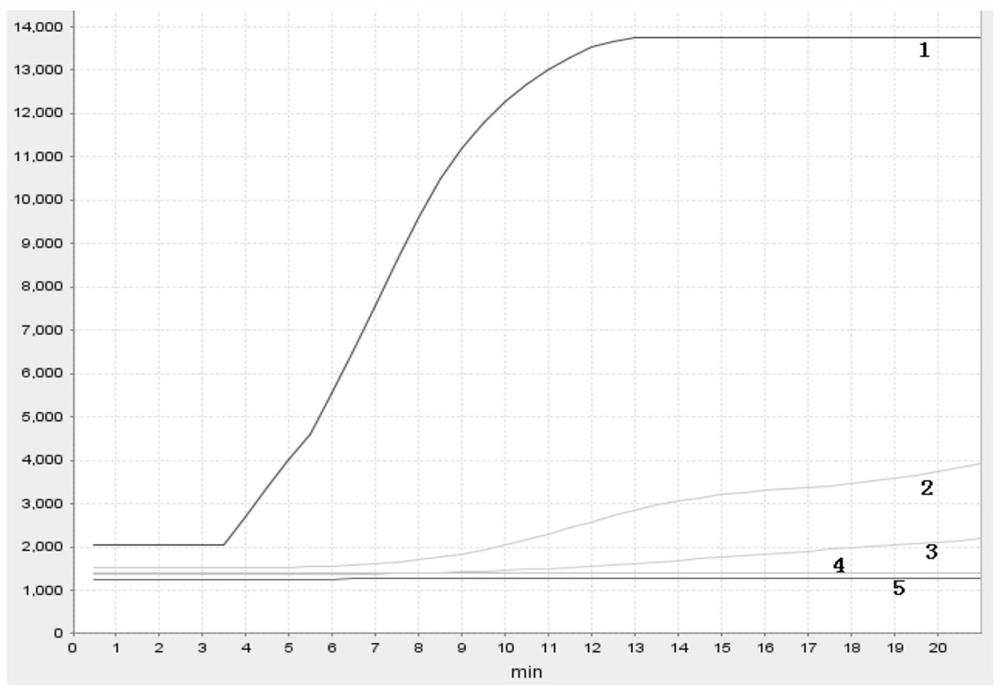
[0121] Select the primer pair and probe sequence designed in Example 2, utilize the recombinase polymerase amplification (in conjunction with endonuclease IV) method amplification reaction system to amplify, construct 50 μ l amplification reaction system as follows:
[0122] 60mM tris-acetate buffer pH8.0
[0123] 100mM potassium acetate
[0124] 14mM magnesium acetate
[0125] 3mM Dithiothreitol
[0126] 5% polyethylene glycol (molecular weight 20000)
[0127] 2mM ATP
[0128] 20mM creatine phosphate
[0129] 100ng / μl creatine kinase
[0130] 400ng / μl E. coli recA protein
[0131] 200ng / μl E. coli SSB protein
[0132] 60ng / μl E. coli recO protein
[0133] 40ng / μl E. coli recR protein
[0134] 60ng / μl E. coli recF protein
[0135] 8Units Bacillus subtilis DNA polymerase I
[0136] 50ng / μl Endonuclease IV
[0137] 450 μM dNTPs
[0138] 420nM per upstream primer
[0139] 420nM each downstream primer
[0140] 120nM fluorescent probe
[0141] The template is a synt...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com