Engineered fc
A technology of antigen-binding molecules and amino acids, which is applied in the field of molecular biology and can solve problems such as contamination of antibody preparations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0307] Example 1: Preparation of Antigen Binding Molecules Comprising Engineered Fc Regions
[0308] The present inventors prepared antigen-binding molecules comprising heavy chains including amino acid substitutions at positions in the CH2 and / or CH3 regions to study the consequences of the substitutions on Fc effector functions.
[0309] An antigen-binding molecule is prepared comprising: (i) a light chain comprising a light chain variable region (VL) and a constant region light chain (CK) of an antibody specific to HER3, and (ii) a heavy chain comprising an antibody specific to HER3 Chain variable region (VH) and human immunoglobulin G1 (G1m3 allotype) heavy chain constant region 1 (CH1), hinge region, heavy chain constant region 2 (CH2) and heavy chain constant region 3 (CH3).
[0310] The CH2 and CH3 regions are either unsubstituted or have the following substitution combinations:
[0311]
[0312] Antigen-binding molecules were expressed using 1) Expi293 Transient ...
Embodiment 2
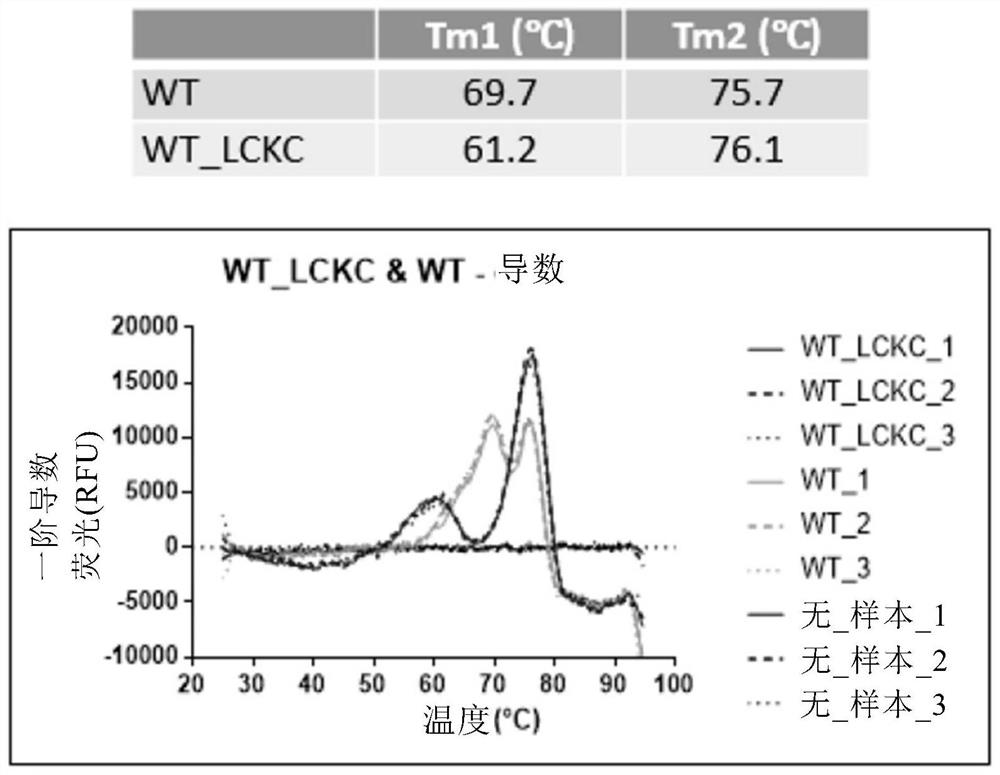
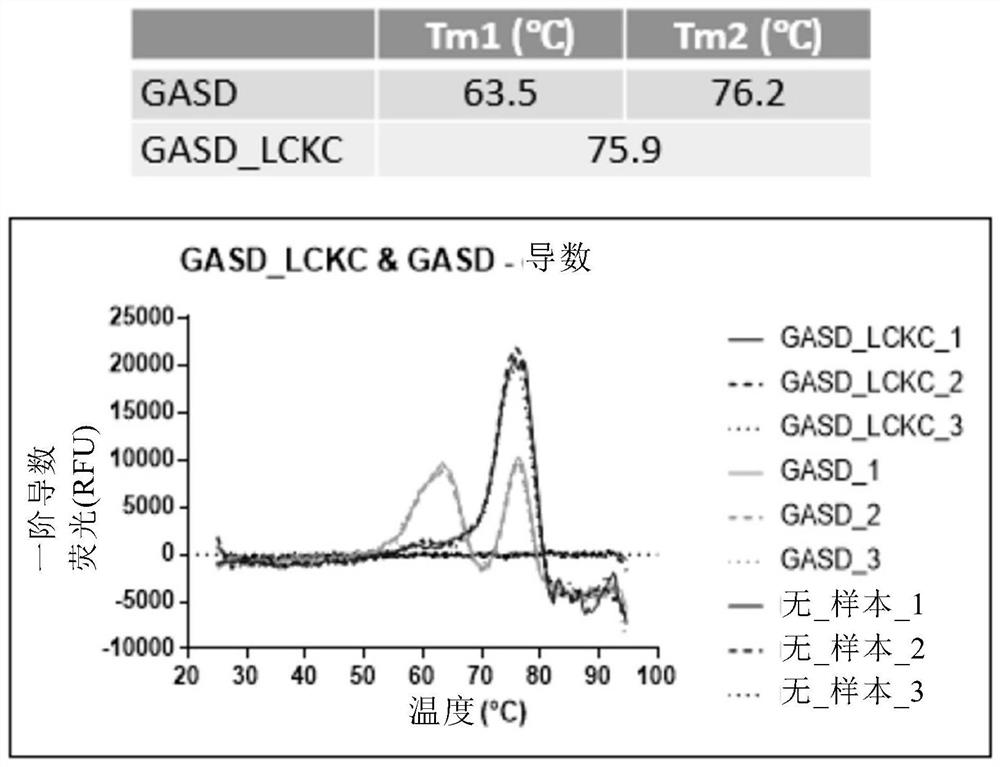
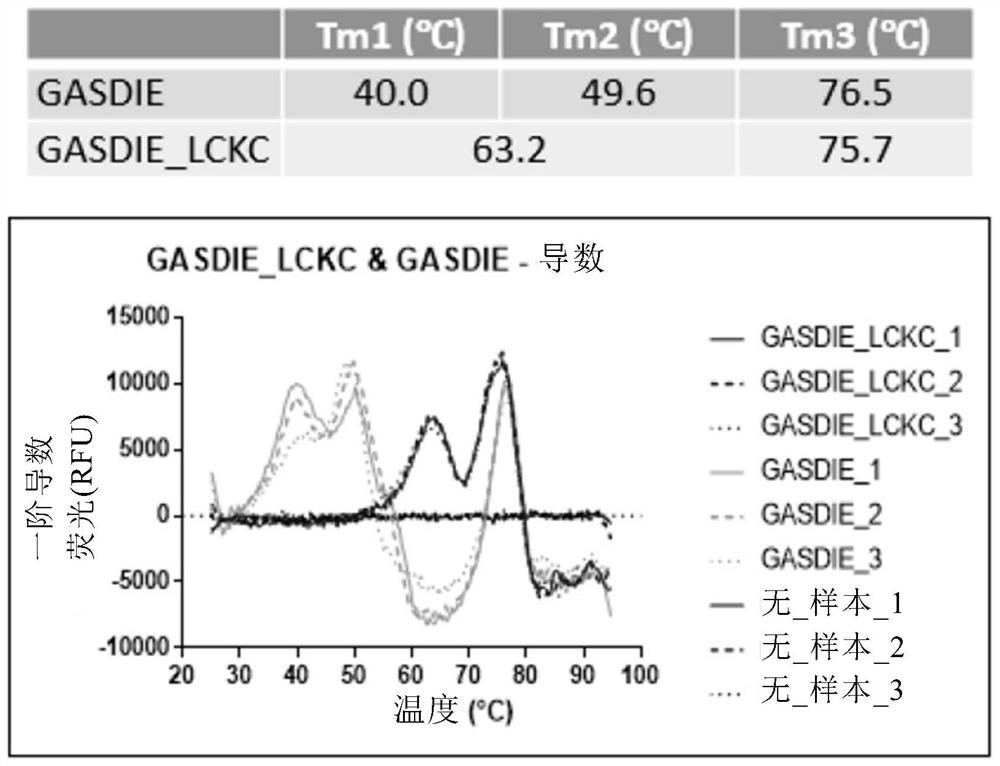
[0325] Example 2: Thermal Stability Analysis of Antigen Binding Molecules Comprising Engineered Fc Regions by Differential Scanning Fluorescence analysis
[0326] The thermal stability of the antigen-binding molecules prepared as described in Example 1 was evaluated by differential scanning fluorescence.
[0327] Briefly, reaction mixtures of 0.2 mg / ml of antibody and Sypro orange dye (Thermo Fisher) were prepared in 25 μl PBS in triplicate and transferred to MicroAmp Optical 96-well reaction plates (Thermo Fisher). wells and sealed with MicroAmp Optical Adhesive Film (Thermo Fisher). Melting curves were run on a 7500 Fast Real-Time PCR System (Applied Biosystems) with TAMRA selected as the reporter gene and ROX as the reference fluorescence. The thermal profile included an initial step of 2 minutes at 25°C and a final step of 2 minutes at 99°C with a ramp rate of 1.2%. Plot the first derivative of the raw data as a function of temperature to obtain a derivative melting ...
Embodiment 3
[0337] Example 3: Antigen-binding molecules containing an engineered Fc region directed against the human Fc receptor FcγRIIIA-158V Affinity analysis
[0338] Antigen binding molecules prepared as described in Example 1 were evaluated for binding to the human Fc receptor FcyRIIa by biolayer interferometry (BLI) using the Pall ForteBio Octet Red384 system.
[0339] Anti-penta-his (His1k) biosensors were purchased from Forte Bio (18-5120) and incubated in PBS buffer (pH 7.2) for 60 s to obtain the first baseline, followed by grouping in PBS pH 7.2 Amino acid-tagged human FcγRIIIa-158V was loaded for 120 seconds. After loading, the biosensor was incubated in PBS buffer (pH 7.2) for 60 seconds to obtain a second baseline, and then the antigen-binding molecules were tested in a diluted series (at concentrations ranging from 15.6 nM to 500 nM) in PBS, pH 7.2. ) for 60 seconds to obtain an association curve. Finally, the biosensor was incubated in PBS pH 7.2 for 120 s to obtain...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com