Non-invasive cancer early screening system based on cfDNA omics characteristics
A cancer and omics technology, applied in the fields of genomics, biochemical equipment and methods, combinatorial chemistry, etc., can solve the problems of easily missed areas of cancer gene copy number variation, not involving specific applications, and lack of comprehensive evaluation of patient cfDNA genomes, etc. Achieving the effect of avoiding omission, large heterogeneity, lower sequencing cost, and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
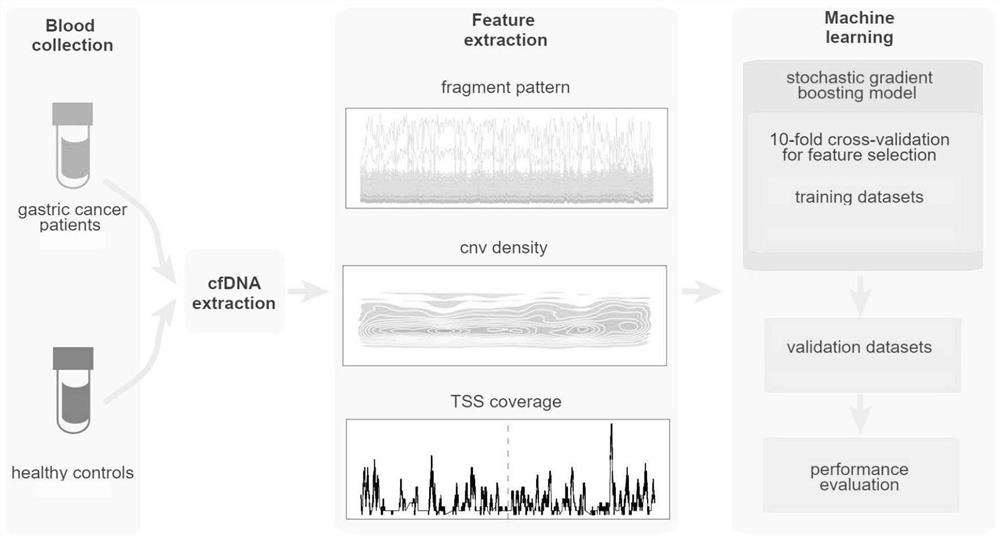
[0067] A noninvasive early screening system for cancer based on cfDNA omics characteristics, including a cfDNA omics characteristic model and a machine learning training model, characterized in that the noninvasive screening method for cancer comprises the following steps:
[0068] S101. Establishing a cfDNA omics feature model;
[0069] S102, blood collection;
[0070] S103, extract cfDNA;
[0071] S104. Building and sequencing the extracted cfDNA;
[0072] S105. Extracting cfDNA omics features and using them for comparison.
[0073] Preferably, the establishment of the cfDNA omics feature model described in step S101 includes the following steps:
[0074] S201. Blood collection;
[0075] S202, extract cfDNA;
[0076] S203. Building and sequencing the extracted cfDNA;
[0077] S204, cfDNA omics feature extraction;
[0078] S205. Machine learning training model.
[0079] Preferably, the blood collection in step S102 and step S201 uses Streck blood collection tubes for ...
Embodiment 2
[0107] This embodiment is carried out on the basis of the above-mentioned embodiment 1, and the similarities with the above-mentioned embodiment 1 will not be repeated.
[0108] This embodiment introduces a method for cfDNA extraction, the specific steps are as follows:
[0109] S601, put the Streck tube in a centrifuge at 4°C, 2000rpm, centrifuge for 10min, and separate the plasma;
[0110] S602. Add 500ul proteinase K and 4ML ACL buffer to the 50ML centrifuge tube containing plasma, mix thoroughly with vortex for 30 seconds, and incubate in a 60°C water bath for 30 minutes;
[0111] S603. Use a vacuum pump to filter the incubated collection tube for 10 minutes, add 600ul of ACW1, 750ul of ACW2, and 750ul of 100% ethanol in order to wash away impurities;
[0112] S604, centrifuge at 12000rpm, centrifuge for 3min;
[0113] S605. Place the collection tube in a metal bath at 50°C for 10 minutes, volatilize ethanol, add 110ul AVE into the collection tube, and incubate at room t...
Embodiment 3
[0116] This embodiment is carried out on the basis of the above-mentioned embodiment 1, and the similarities with the above-mentioned embodiment 1 will not be repeated.
[0117] This embodiment introduces a method for feature extraction of cfDNA three-omics, including the following steps:
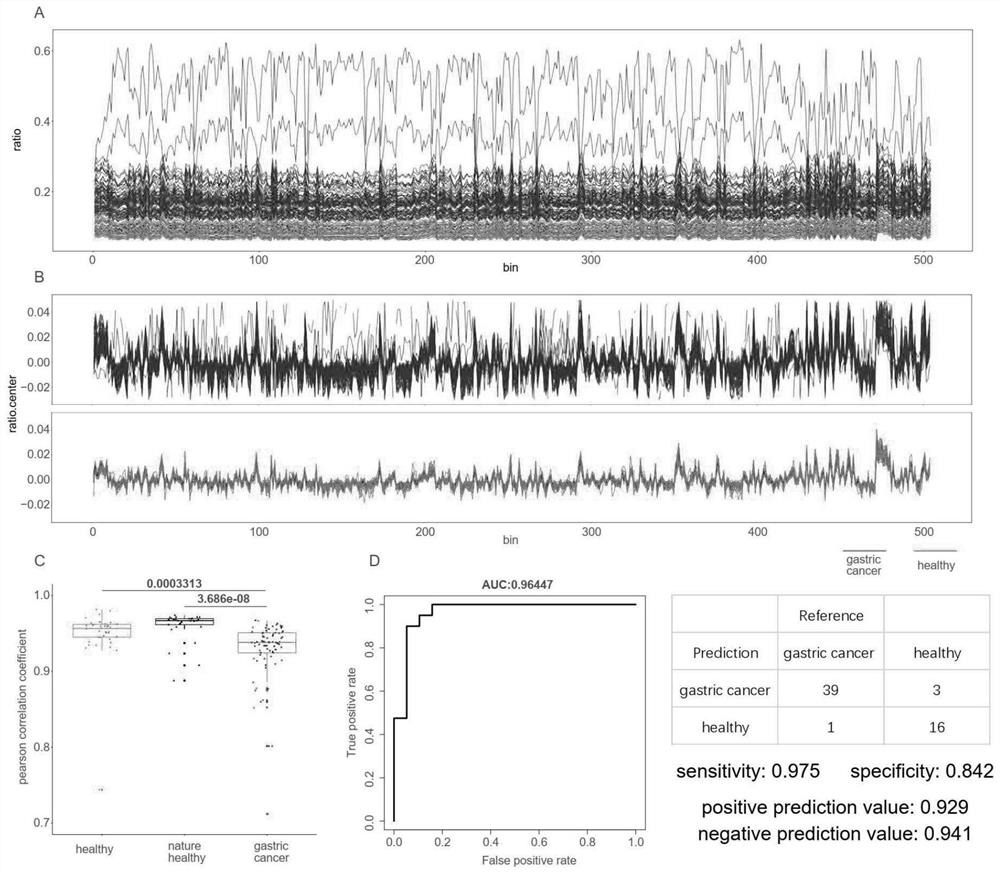
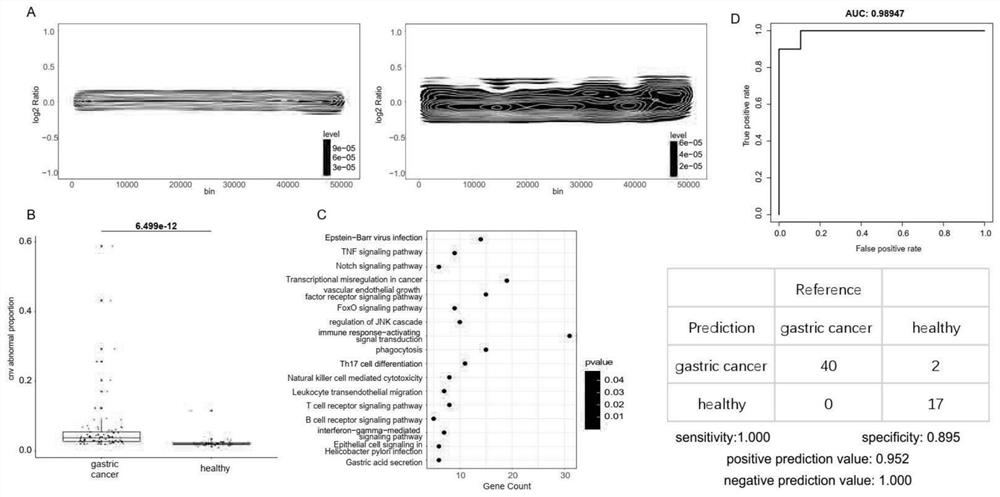
[0118] A.fragment pattern: First compare the cfDNA sequence file to the reference genome hg19, discard low-quality sequences and filter out repeated sequences from the resulting BAM file; then compare the low-coverage regions of the hg19 reference genome and the Duke black box The region is excluded; next, the hg19 autosome is divided into 504 contiguous and non-intersecting fragments, each fragment is 5mb in length; the number of cfDNA with a length greater than 150bp and the number of cfDNA with a length less than 150bp are counted in each fragment region ; Then use the LOESS regression method to correct the GC content of these cfDNA numbers, and then use the mean normalization method to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com