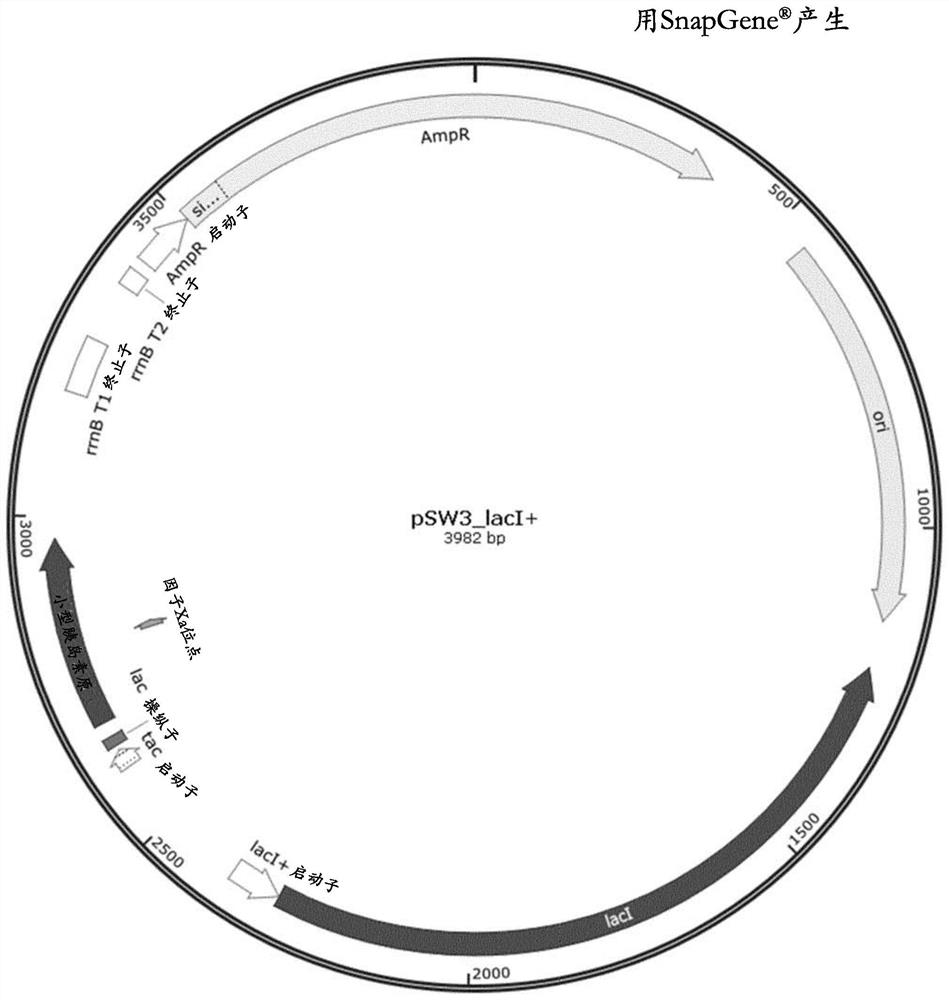
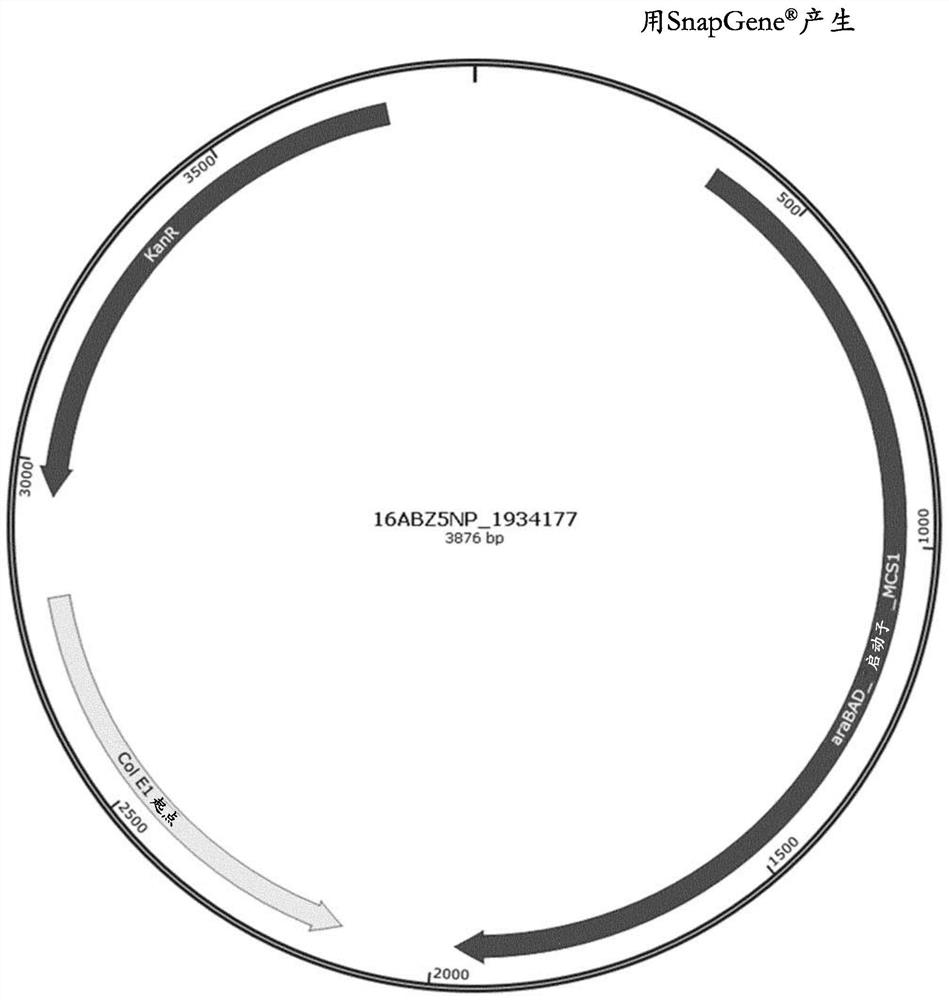
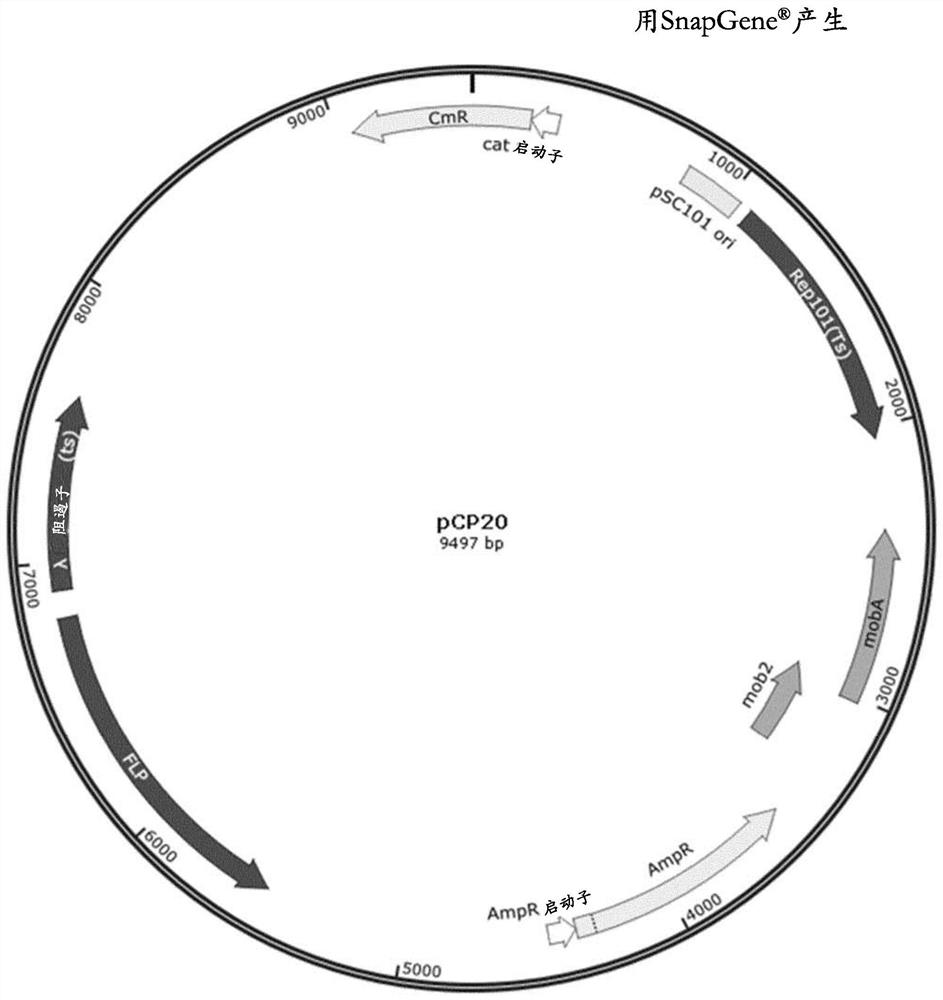
Method for reducing misincorporation of non-canonical branched-chain amino acids
A branched-chain amino acid, non-classical technology, applied in the field of reducing the mis-incorporation of non-classical branched-chain amino acids, can solve the problem of incomplete clarity of non-classical branched-chain amino acids and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] As noted above, the present invention relates to methods for producing recombinant polypeptides of interest in microbial host cells. The method comprises (a) introducing into a microbial host cell a polynucleotide encoding the polypeptide of interest that has been modified such that an enzyme activity as described elsewhere herein in the microbial host cell is compatible with said enzymatic activity is modulated in an unmodified microbial host cell; and (b) expressing said polypeptide of interest in said microbial host cell.
[0028] According to the present invention, the recombinant polypeptide of interest should be produced in a microbial host cell. The term "recombinant polypeptide" as used herein refers to a genetically engineered polypeptide. Therefore, the polypeptide to be produced should be heterologous with respect to the microbial host cell, meaning that the host cell does not naturally express the polypeptide of interest. Thus, the term "heterologous" mean...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com