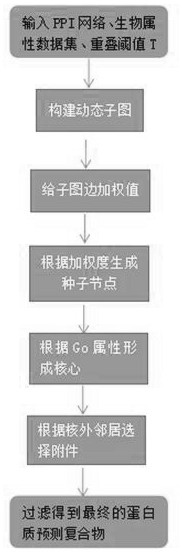
Protein complex identification method based on core-accessory structure
A protein complex and identification method technology, which is applied in the field of identifying protein complexes by integrating network topology information and protein biological properties, can solve the problems of false positives in protein interaction data, ignoring protein biological features, and noise in PPI networks. Achieve the effect of expanding application range and practicability, overcoming high data noise, and improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0111] We tested our proposed algorithm BOCAS on three datasets of Gavin, Krogan, and DIP respectively. Table 1 gives the detailed information of Gavin, Krogan, and DIP three data sets, including the number of proteins contained in each network and the number of interactions between proteins. Table 2 presents the information of the protein biological attribute dataset.
[0112] Table 1 Protein Interaction Network Dataset
[0113] PPI dataset protein quantity number of interactions Gavin 1430 6531 Krogan 2674 7075 DIP 5093 24743
[0114] Table 2 Protein biological attribute data set
[0115] biological data set Remark Gene Expression Profiles Version: GSE3431, includes expression values for 36 time points per gene Go annotation set Go annotations include Go annotation information for 7014 proteins known complex set CYC2008
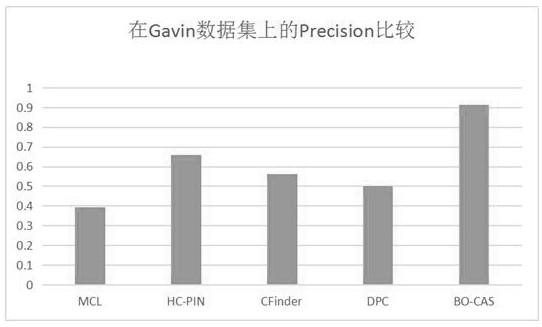
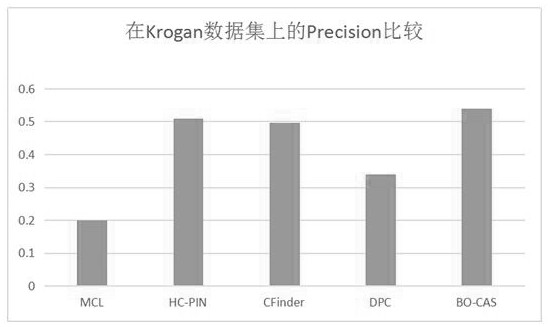
[0116] To evaluate the performance of the BO-CAS method in protein complex...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com