Protein ubiquitination modification site detection method based on high-precision mass spectrum and application
A detection method and ubiquitin-like technology, which is applied in the detection and application of protein ubiquitin-like modification sites, can solve problems such as difficulty in collecting fragment ion information, low ionization efficiency, and inability to truly reflect the modification level. The effect of cost reduction and high-throughput quantitative analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
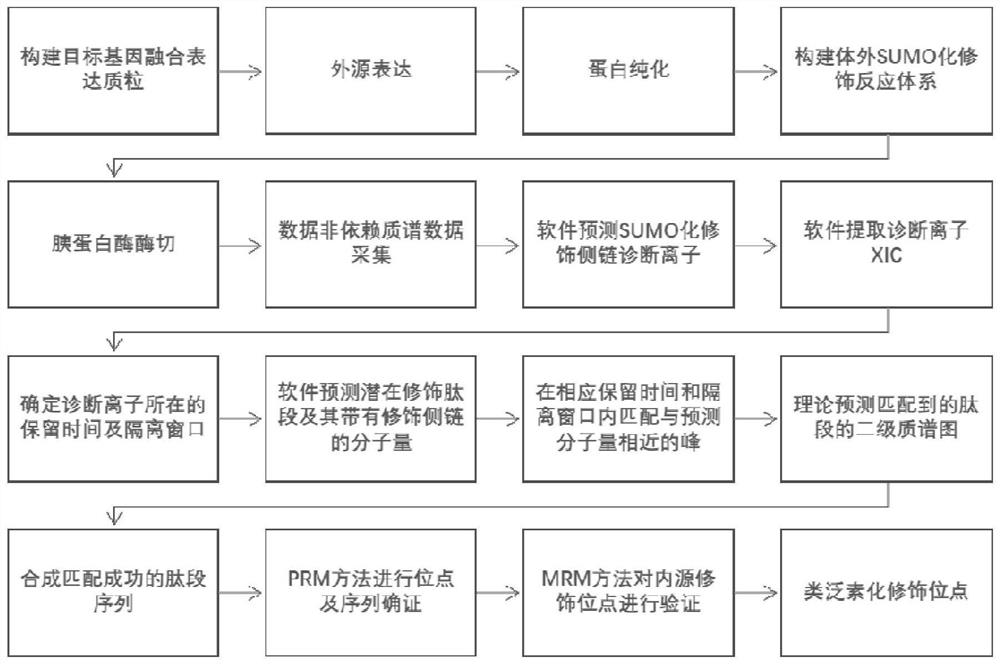
[0068] figure 1 Schematic diagram of the identification process for SUMO modification sites. Such as figure 1 As shown, this embodiment provides a method for detecting protein ubiquitination modification sites, which comprises the following steps:
[0069] (1) For the target protein to be studied, construct the target gene fusion expression plasmid through gene cloning technology;
[0070] (2) Transfer the fusion expression plasmid constructed in step (1) into an exogenous gene expression system for exogenous expression;
[0071] (3) Purifying the fusion protein expressed in step (2) by affinity chromatography;
[0072] (4) Constructing an in vitro SUMOylation modification reaction system;
[0073] (5) Trypsin digestion of the protein in the reaction system;
[0074] (6) The mass spectrum data of the polypeptide obtained in step (5) of liquid chromatography mass spectrometry tandem collection;
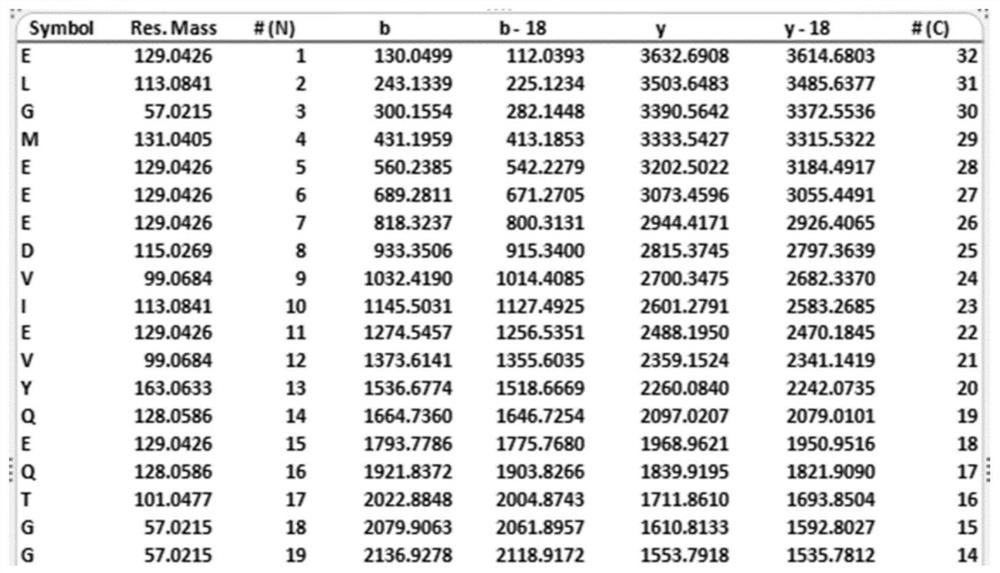
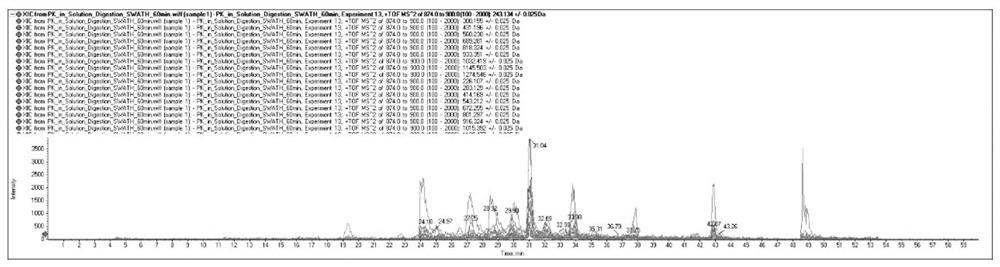
[0075] (7) Mass spectrometry data analysis to obtain the sequence of modific...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com