Single-cell RNA-seq data clustering method based on deep noise reduction auto-encoder
An autoencoder and data clustering technology, applied in the field of single-cell RNA-seq data analysis, which can solve problems such as inability to cluster data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0025] The present invention is described in further detail below in conjunction with embodiment.
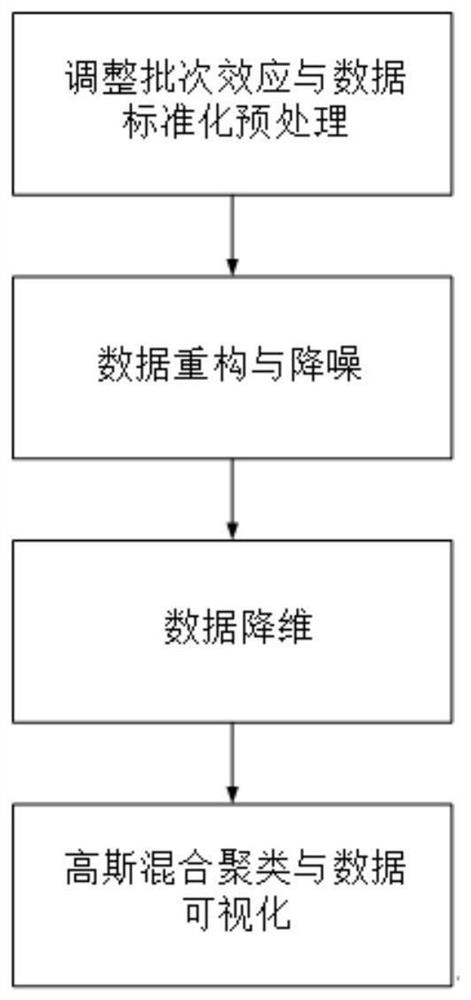
[0026] Such as figure 1 As shown, it shows the four steps of the present invention to improve the clustering effect of single-cell RNA-seq data based on the deep noise reduction self-encoder, adjusting the batch effect and data standardization preprocessing, data reconstruction and noise reduction, data dimensionality reduction, Gaussian mixture clustering and data visualization.
[0027] The invention provides a method for clustering single-cell RNA-seq data based on a deep layer noise reduction autoencoder. The invention comprises the following steps:
[0028]Step 1. Adjust the batch effect and data standardization preprocessing. The present invention uses 5 public data sets downloaded from ArrayExpress and GEO databases to verify the effectiveness of the present invention. The gene expression values in these 5 public data sets are taken from various tissue cells, includin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com