A nucleic acid-based method for tree phenotype prediction
A phenotypic and nucleic acid technology, applied in horticultural methods, botanical equipment and methods, and microbial determination/inspection, etc., can solve the problem of not giving the degree of hybridization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
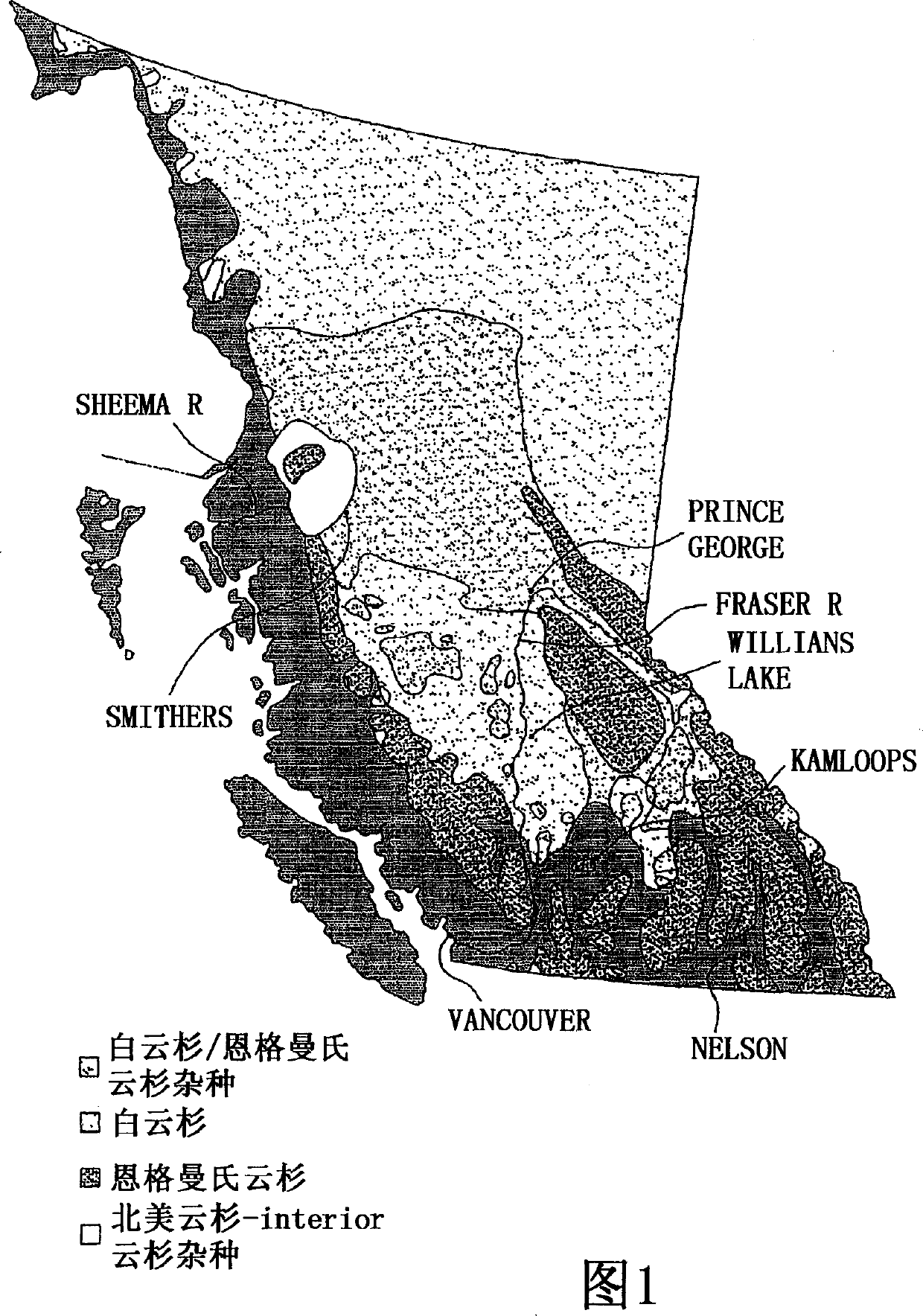
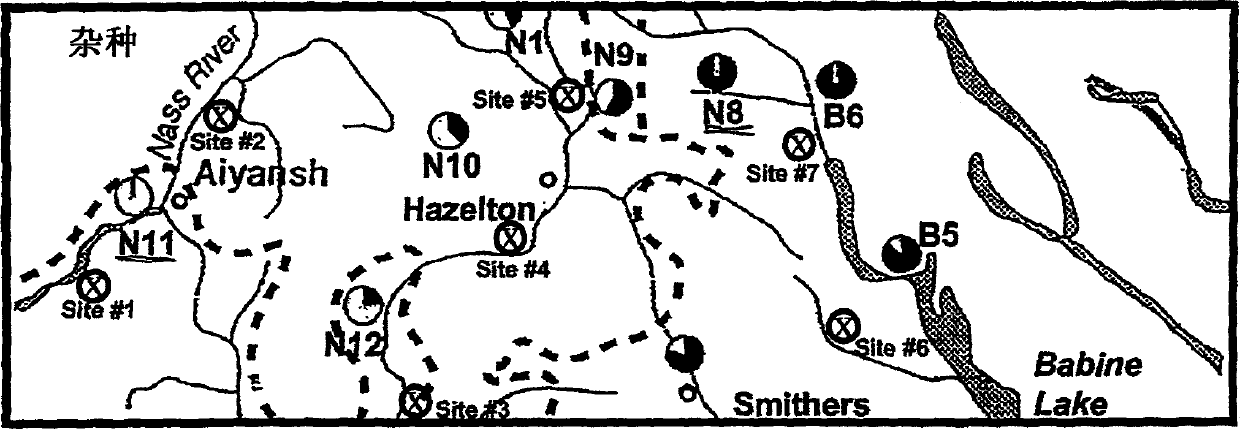
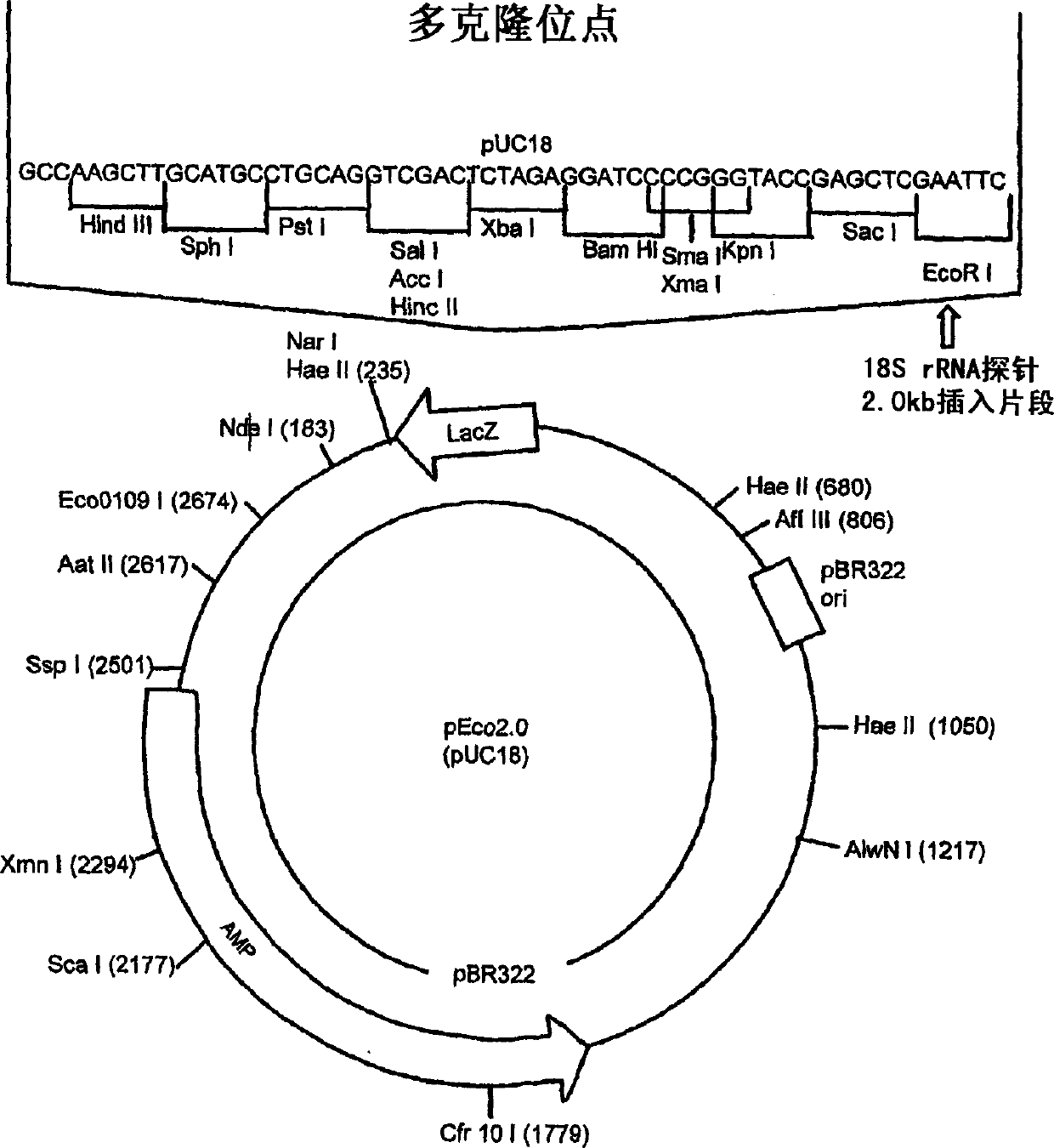
[0064] The present invention provides a novel method for predicting selected phenotypes and rapidly selecting superior trees for known pulp and paper production lines using DNA probes. The method comprises the steps of isolating tree genome DNA from spruce living tissue, hybridizing the genome DNA with a DNA probe, and evaluating the intensity of the hybridization pattern obtained by optical density.
[0065] A particular embodiment of the present invention is to determine the exact extent of genetic admixture (or introgression) of two parental species in a hybrid population. Due to - for example in hybrid spruce populations - the linear relationship between the degree of genetic introgression and fiber length (for the purposes of the present invention), the strength of the DNA probe hybridization profile can be used to directly, precisely and reproducibly Predict the fiber length of individual Picea spruce / interoir spruce hybrids (for a given tree age) in a population.
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com