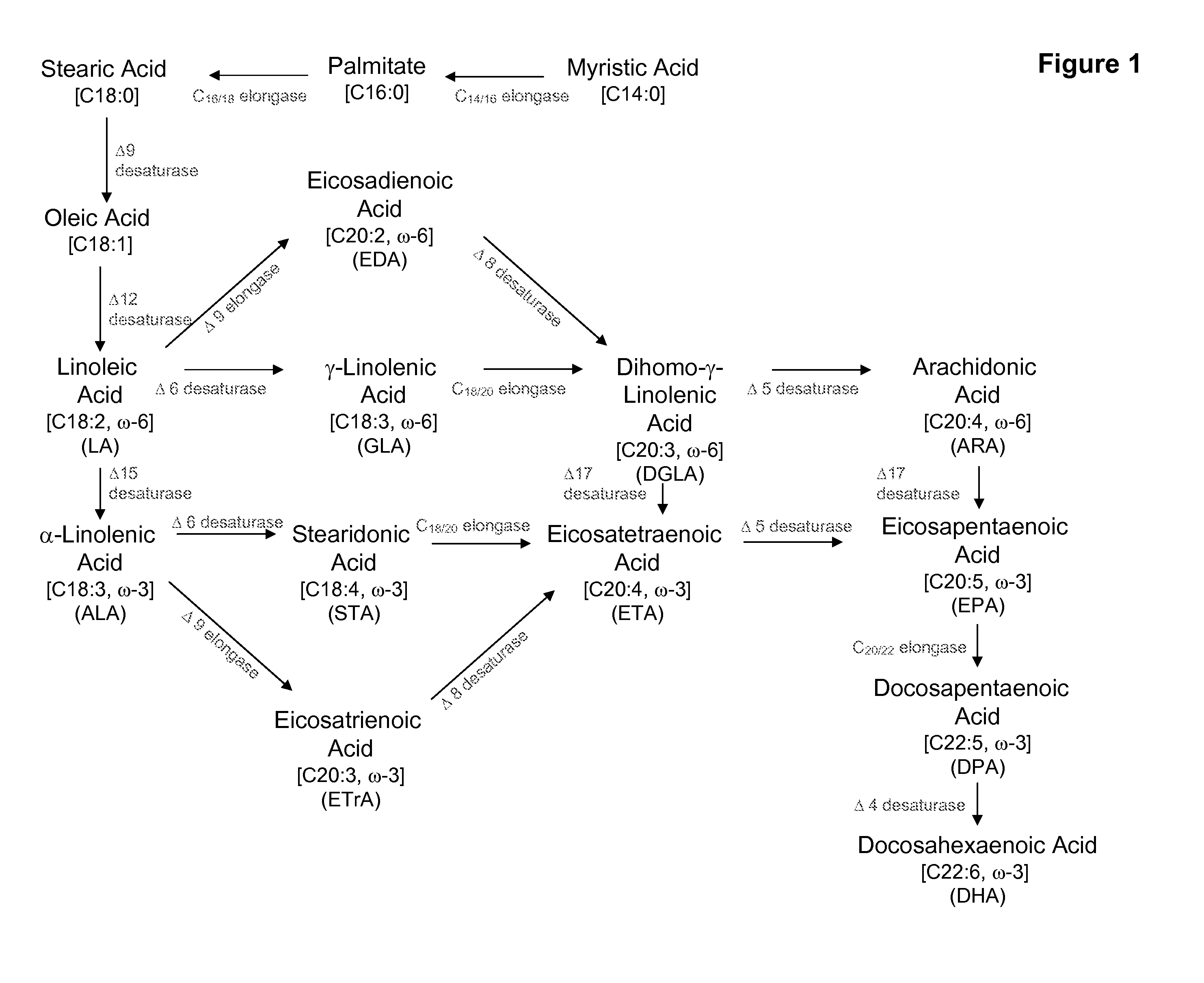
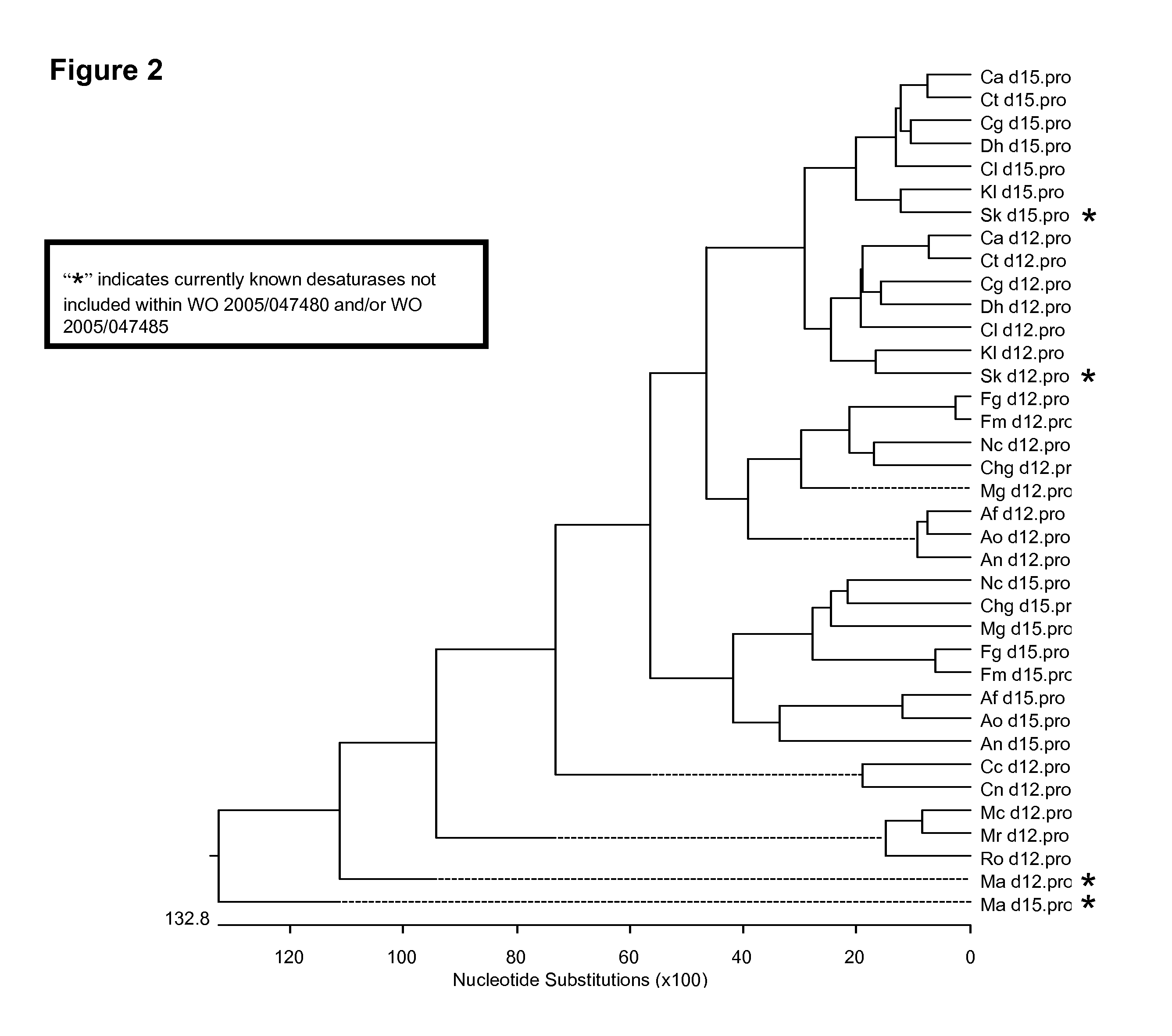
Fungal delta 12 desaturase and delta 15 desaturase motifs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Description Of Δ12 Knockout Yarrowia Strain L38
[0266]Strain L38 is a Δ12 desaturase-disrupted strain of Yarrowia lipolytica similar to the d12KO strain described as Q-d12D (PCT Publication No. WO 2004 / 10416). It was also derived from Y. lipolytica ATCC #76982 following disruption of its only native Δ12 desaturase gene by replacement with a disrupted version via homologous recombination.
Selection Method Theory
[0267]The methodology used to create the d12KO strain identified herein as L38 relied on site-specific recombinase systems. Briefly, the site-specific recombination system consists of two elements: (1) a recombination site having a characteristic DNA sequence [e.g., LoxP]; and, (2) a recombinase enzyme that binds to the DNA sequence specifically and catalyzes recombination (i.e., excision) between DNA sequences when two or more of the recombination sites are oriented in the same direction at a given interval on the same DNA molecule [e.g., Cre]. For the purposes herein, an integ...
example 2
Identification Of Fungal Sequences Encoding Δ15 Desaturases
[0274]“Pairs” of Δ12 / Δ15 desaturase-like polypeptides have previously been identified in the following filamentous fungi: Fusarium moniliforme, Fusarium graminearum, Magneporthe grisea, Neurospora crassa, Aspergillus nidulans, Mortirerella alpina and Saccharomyces kluveromyces. In each case, one protein was subsequently determined (or predicted) to possess Δ12 desaturase activity while the other protein was determined to possess Δ15 desaturase activity (PCT Publication No. WO 2005 / 047480; PCT Publication No. WO 2005 / 047485; Sakuradani et al., Appl. Microbiol. Biotechnol., 66:648-654 (2005); Oura, T. and Kajiwara, S., Microbiology (Reading, Engl.), 150(6):1983-1990 (2004); Sakuradani, E., et al., Eur. J. Biochem., 261(3):812-820 (1999); Watanabe, K., et al., Biosci. Biotechnol. Biochem., 68(3):721-727 (2004)). More specifically, the following proteins were previously characterized as Δ15 desaturases: Fm.d15 (SEQ ID NOs:39 and...
example 3
Amino Acids Motifs For Identification Of Fungal Δ12 And Δ15 Desaturases
[0290]Comparison of the different fungal Δ12 / Δ15 desaturase-like polypeptides (i.e., including both known and putative Δ12 and Δ15 desaturase sequences) from sixteen different fungal species (Table 4, supra) enabled the identification of regions of significant homology between the genes, such as the 15 amino acids surrounding and including the conserved His Box I (“HE[C / A]GH”; SEQ ID NO:6). More specifically, a total of 7 different sequence variants within this region (i.e., from 6 residues upstream of the His Box I to 4 residues downstream of the His Box I) were identified within the Δ12 desaturases (i.e., SEQ ID NOs:7, 8, 9, 10, 11, 12 and 13), while a total of 9 different sequence variants within this region were identified within the Δ15 desaturases (i.e., SEQ ID NOs:22, 23, 24, 25, 26, 27, 28, 29 and 30). These conserved sequences are summarized below in Tables 12 and 13 (wherein the shaded portion of the se...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com