Data processing, analysis method of gene expression data to identify endogenous reference genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Gene Expression Dataset Construction
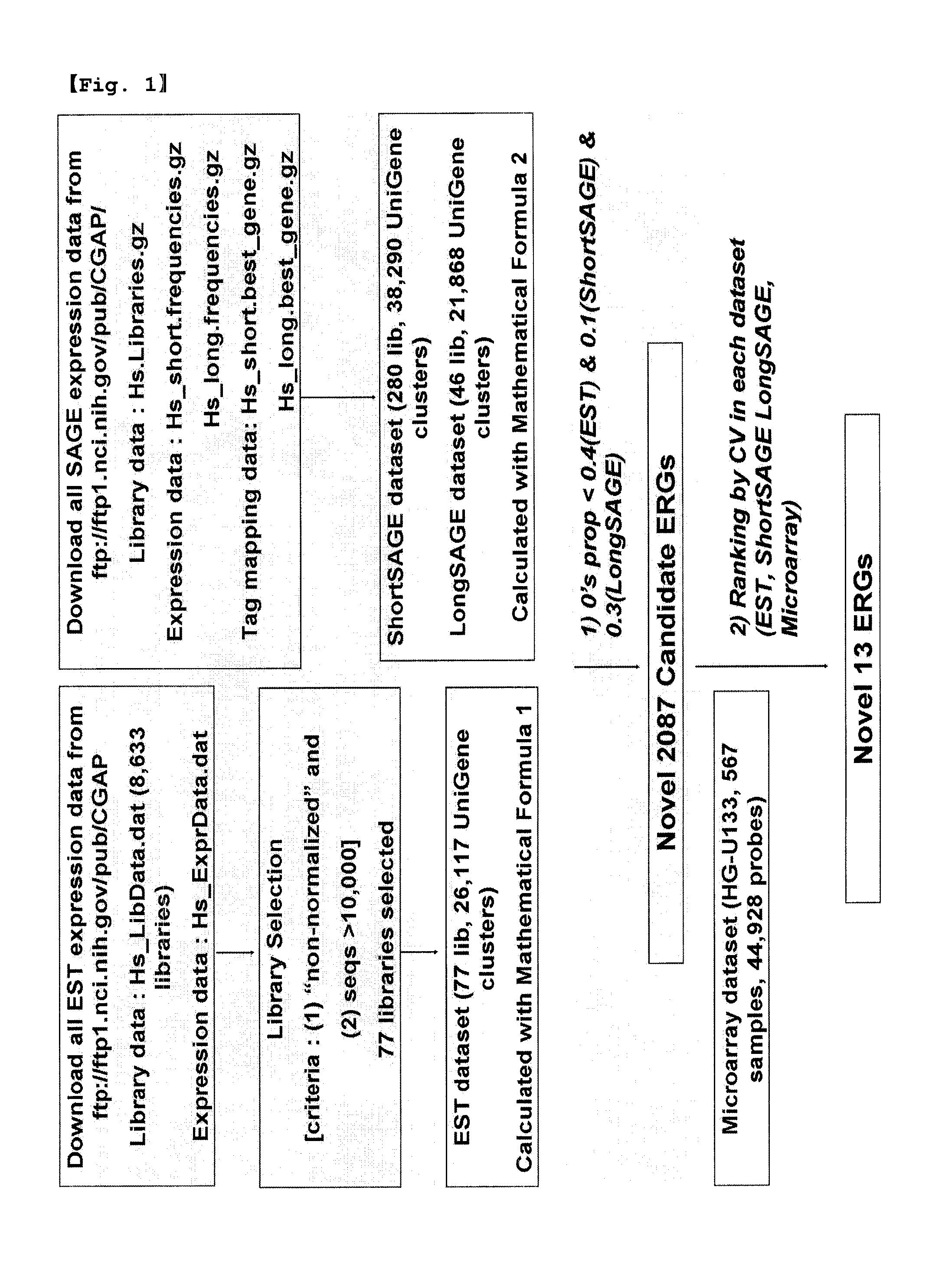
[0075]EST (expressed sequence tag) and SAGE (serial analysis of gene expression) human gene expression data were collected from the publicly available CGAP site (The Cancer Genome Project, http: / / cgap.nci.nih.gov / ). Microarray gene expression data were obtained from the GeneExpress Oncology Datasuite™ of Gene Logic Inc., based on the Affymetrix Human Genome U133 array set.
[0076]Out of a total of 8,633 libraries in Hs_LibData.dat (31 / Oct / 05) file, 77 libraries meeting the requirements: 1) non-normalized and 2) >10,000 sequences, were included in the EST dataset constructed from the CGAP site. EST frequency in each library was obtained from Hs_ExprData.dat (31 / Oct / 05) file. 29 different tissues, including normal and tumor samples, and 26,117 UniGene clusters were included in these libraries.
[0077]SAGE short data for all 280 libraries (Hs_short.frequencies.gz, 05 / Dec / 06), representing 38,290 UniGene clusters, and SAGE long data (Hs_long.frequencies.g...
example 2
Selection of Candidate ERG
[0079]Using the datasets constructed in Example 1, expression level was calculated for UniGene clusters so as to search for housekeeping genes which are constitutively expressed in most human tissues.
[0080]Gene expression levels of EST dataset for each gene were calculated as the number of ESTs of a gene in a given library, divided by the total number of ESTs in all genes in a given library and then multiplied by 1,000,000, as expressed by Mathematical Formula 1.
[0081]Likewise, Gene expression levels of SAGE dataset for each gene were calculated as the number of tags (sum of tag frequency) of a gene in a given library, divided by the total number of tags and then multiplied by 1,000,000, as expressed by Mathematical Formula 2.
<MathematicalFormula1>ETSgeneexpression= NoofESTofaGivenGeneinLibraryTotalNo.ofESTsinLibaray×1,000,000<MathematicalFormula2>SAGEgeneexpression=NoofTagsofaGivenGeneinLibraryTotalNo.ofTagsinLibaray×1,000,000
[0082]Expression l...
experimental example 1
Analysis of Candidate ERG
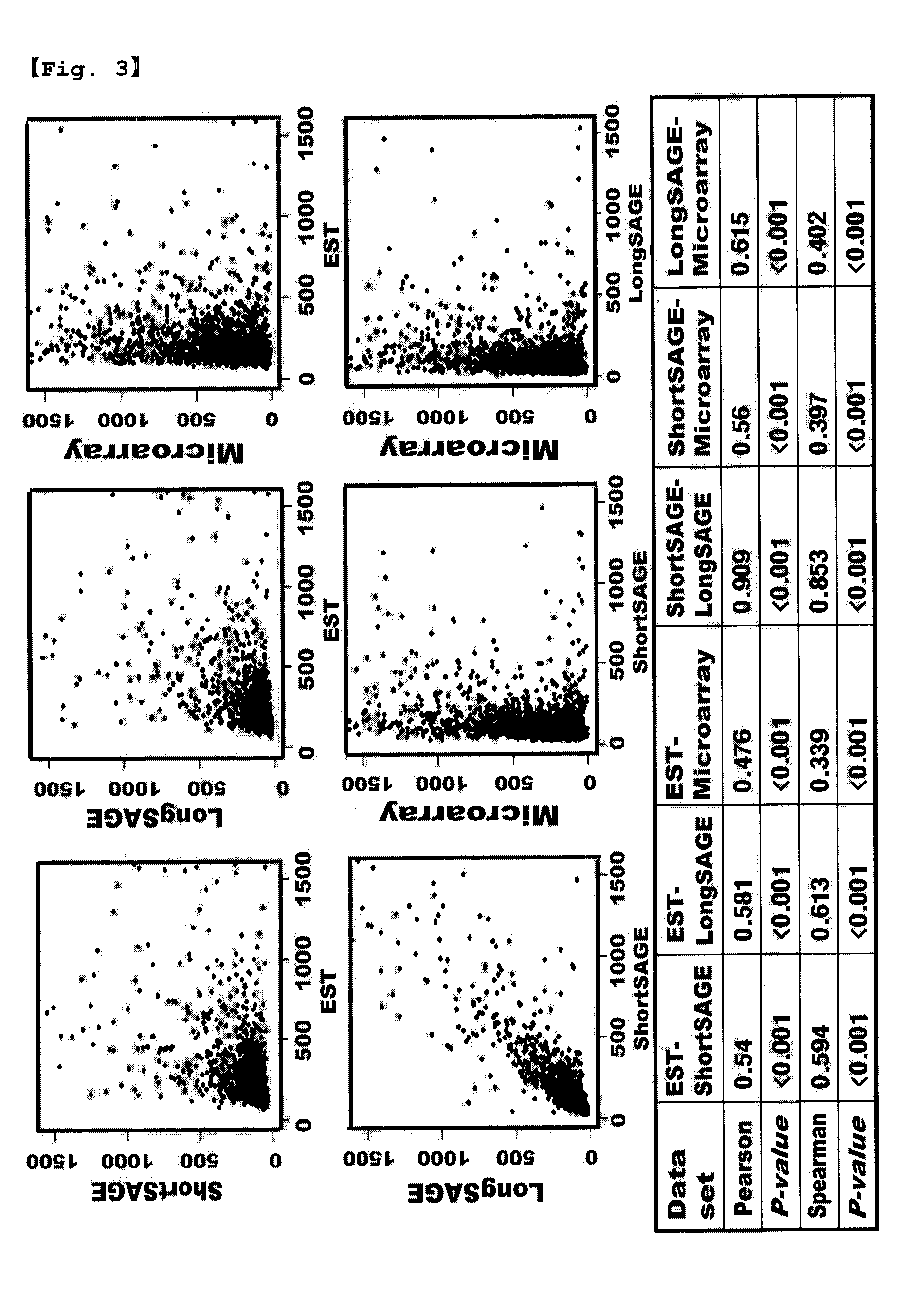
[0089] Functional Classification of Candidate ERG
[0090]Using FunCat (Functional Classification Catalogue, Version 2.0, Ruepp, A., et al. Nucleic Acids Res, 32, 5539-45, 2004), the 2,087 genes obtained above were classified.
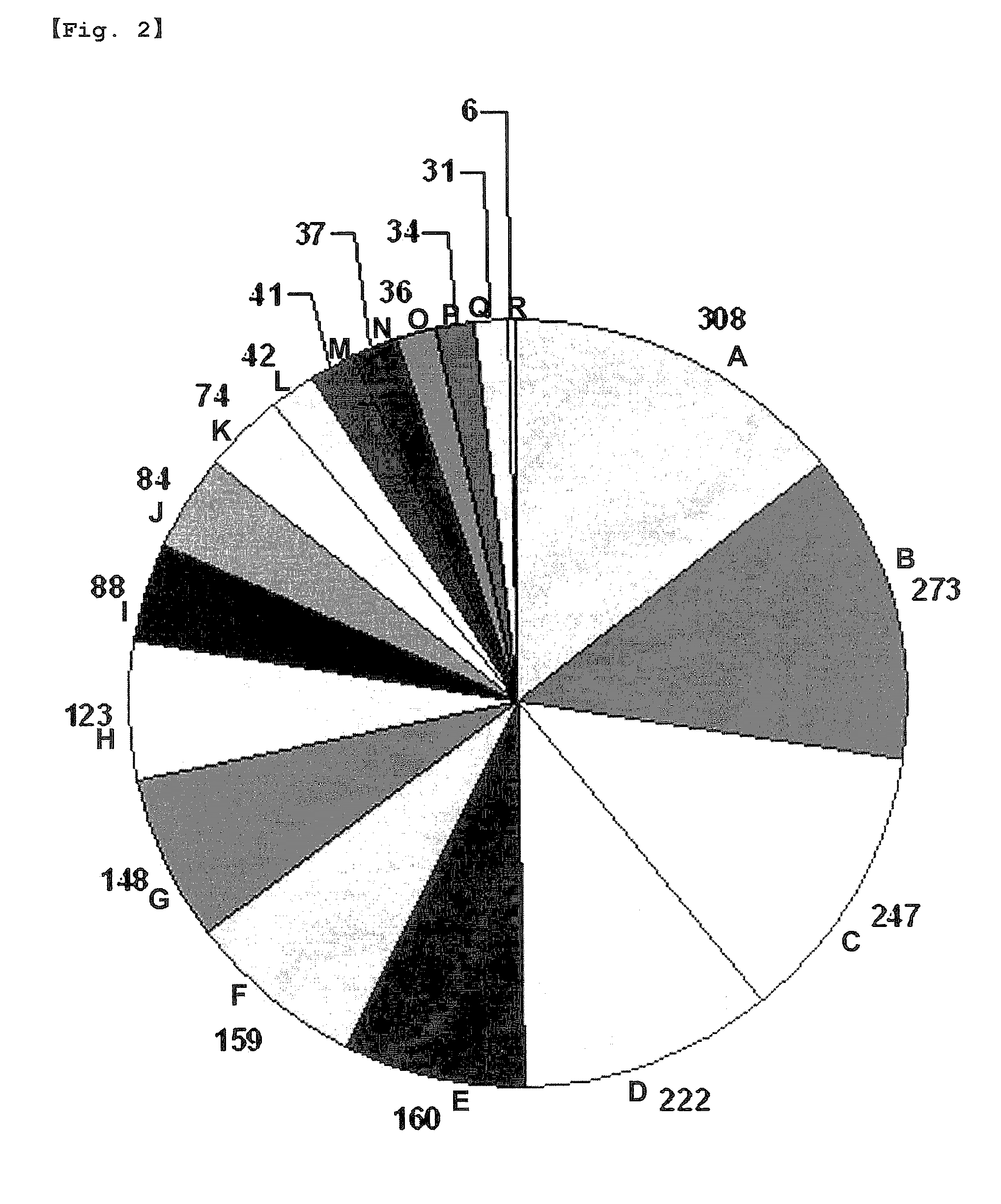
[0091]Out of the 2,087 candidate ERGs, 1,689 UniGene clusters were associated with GO terms allocated to MIPS (Munich Information Center for Protein Sequences) FunCat (Functional Catalogue). Among the 1,689 UniGene clusters, 1,318 UniGene clusters were functionally classified to be associated with GO terms corresponding to biological processes. These 1,318 genes were identified to be involved in various basic cellular functions. While a high proportion of the previously reported traditional HKGs encode metabolism and ribosome proteins (Eisenberg E & Levanon E Y, Trends Genet 19:362-5, 2003; Hsiao L L et al., Physiol Genomics 7:97-104, 2001), genes encoding proteins involved in protein fate (23%, 308 / 1318) and cellular transport (21%, 273 / 1...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Electric charge | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com