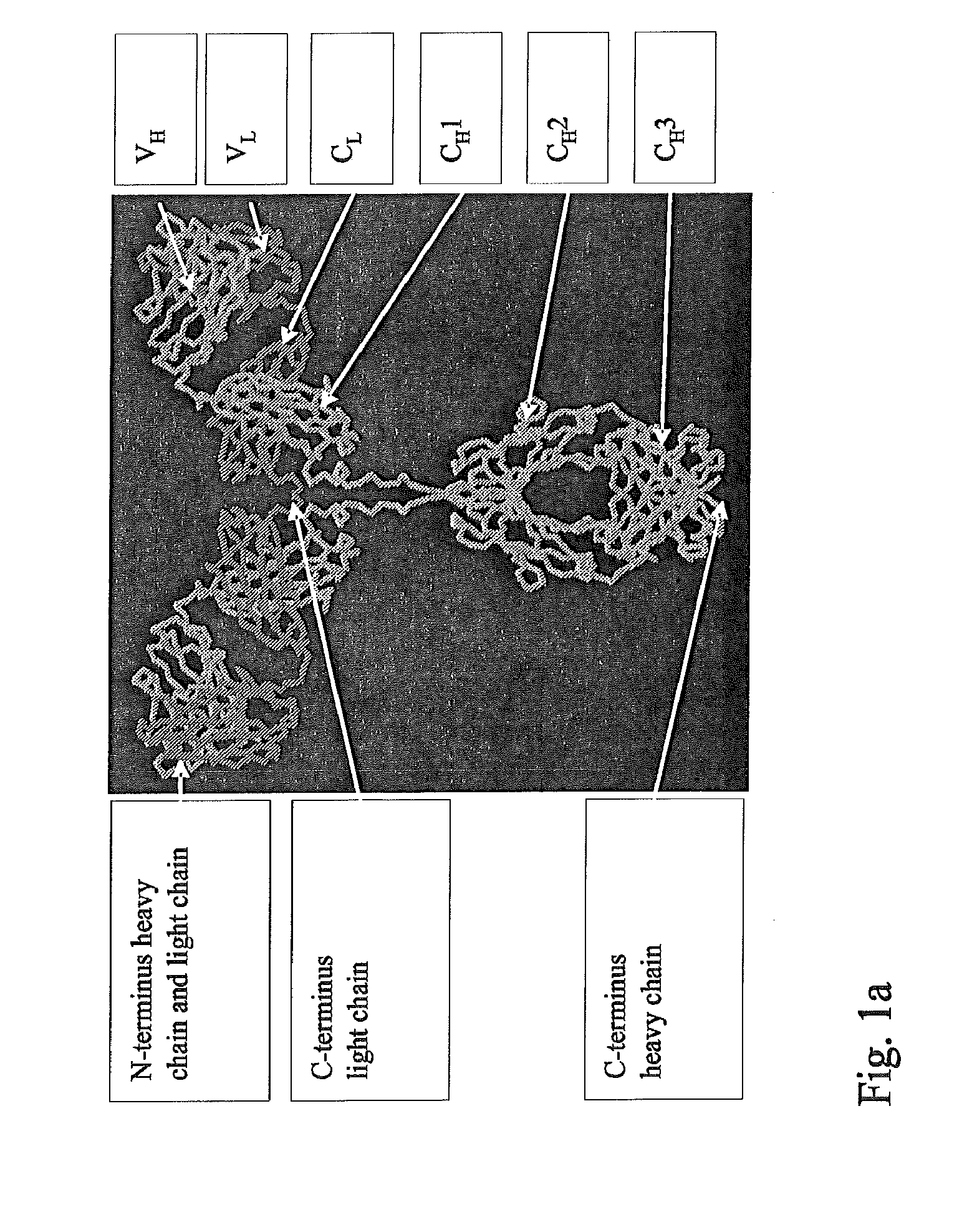
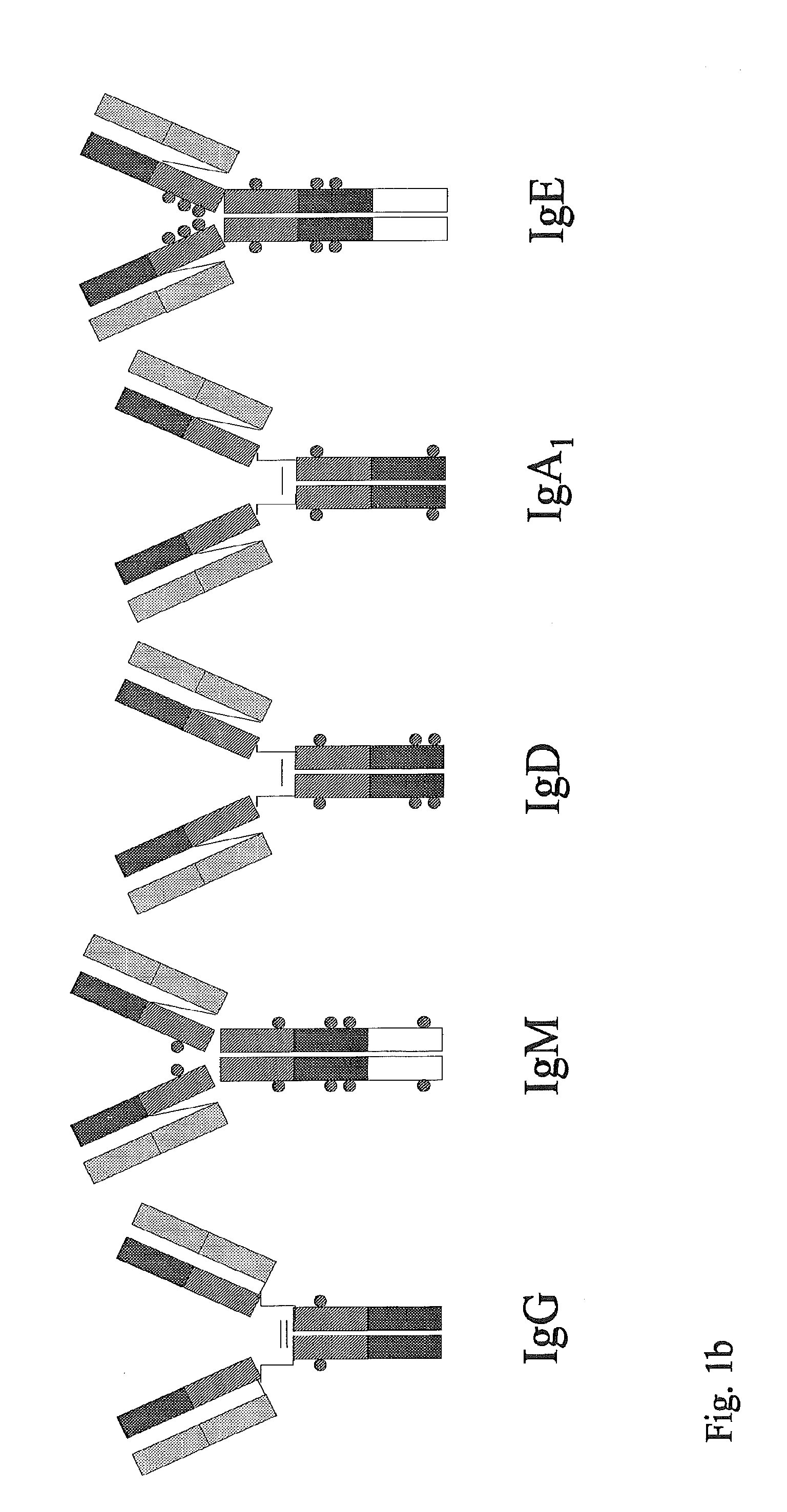
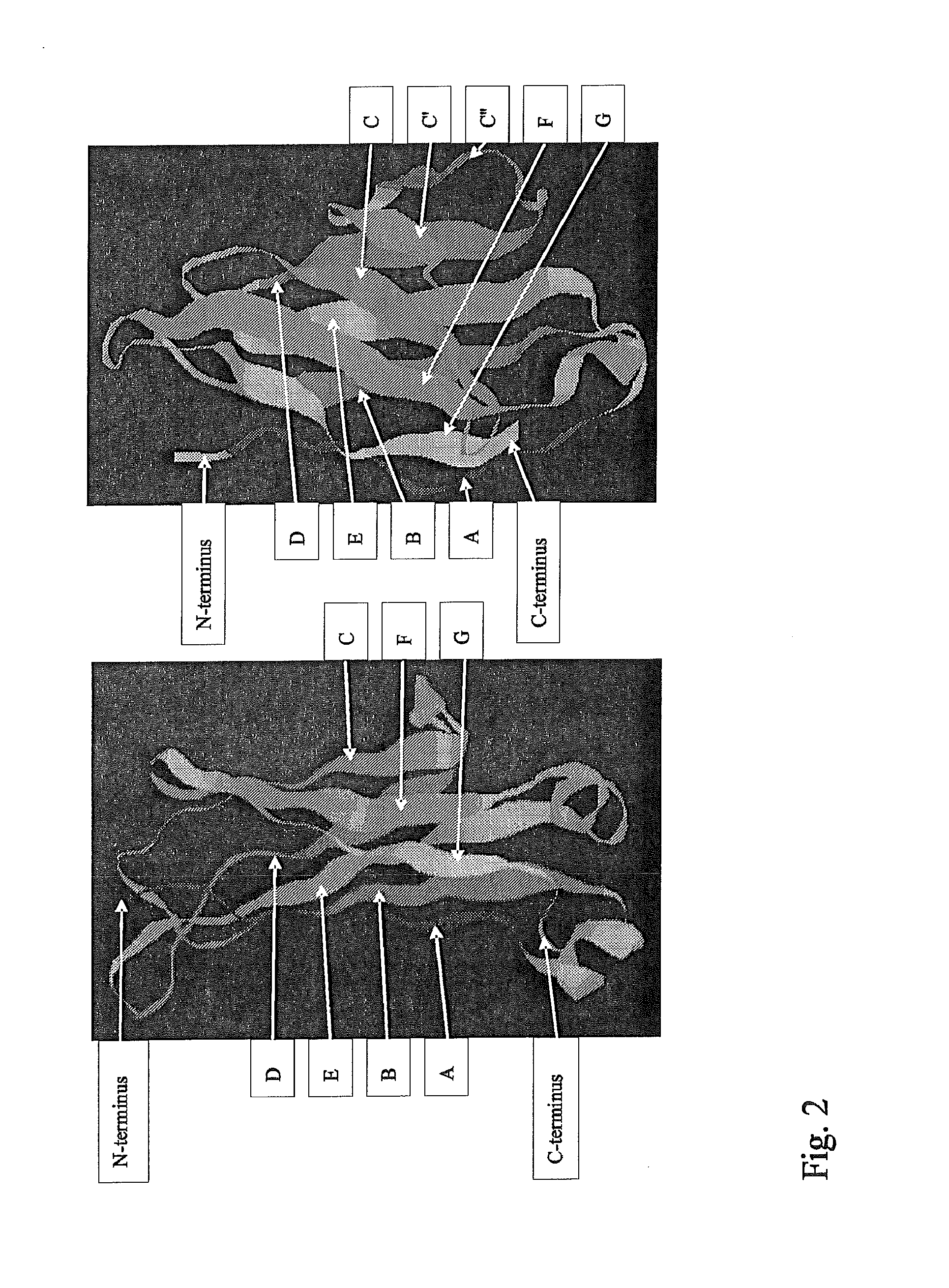
Synthetic immunoglobulin domains with binding properties engineered in regions of the molecule different from the complementarity determining regions
a technology of immunoglobulin and binding properties, applied in the field of libraries of immunoglobulins, can solve the problems of peptide grafting into a non-cdr loop, difficult to further modify the structural loop without changing, and high chance of being inactiv
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
Construction of the CH3 Library and Phage Surface Display
[0252]The crystal structure of an IgG1 Fc fragment, which is published in the Brookhaven Database as entry 1OQO.pdb was used to aid in the design of the mutated CH3 domain.
[0253]The sequence which was used as the basis for construction of the CH3 library is given in SEQ ID No. 1. In this sequence, the first amino acid corresponds to Proline 343 of chain A of Brookhaven database entry 1oqo.pdb. The last residue contained in 1oqo.pdb is Serine 102 of SEQ ID No. 1. After detailed analysis of the structure of 1oqo.pdb and by visual inspection of the residues forming the loops which connect the beta strands, it was decided to randomize residues 17, 18 and 19, which are part of the loop connecting beta strand A-B as well as 71, 72, 73, 76, and 77, which are part of the loop connecting beta strand E-F of SEQ ID No. 1. A molecular model of the engineered CH3 domain, with the randomized part indicated by a solvent accessible surface is...
example 2
Construction of the CH3+3 Library
[0255]This library was constructed and cloned in the same way as the CH3 library. The amino acid sequence of the construct is given in SEQ ID No. 10, the corresponding nucleotide sequence in SEQ ID No. 11, and the primers used for construction were SEQ ID No. 4-7, SEQ ID No. 9 and SEQ ID No. 12.
example 3
Construction of the CH3+5 Library
[0256]This library was constructed and cloned in the same way as the CH3 library. The amino acid sequence of the construct is given in SEQ ID No. 13, the corresponding nucleotide sequence in SEQ ID No. 14, and the primers used for construction were SEQ ID No. 4-7, SEQ ID No. 9 and SEQ ID No. 15.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Volume | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com