Nucleic acid amplification methods
a technology of nucleic acid and amplification method, which is applied in the direction of microorganism testing/measurement, material analysis, biochemistry apparatus and processes, etc., can solve the problems of time and laborious process, unable to achieve general applicability in clinical practice, and unable to achieve sensitive and standardized detection, rapid and sensitive detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Detection of HIV-1 RNA in a Sample
Preparation of Oligonucleotide Probes
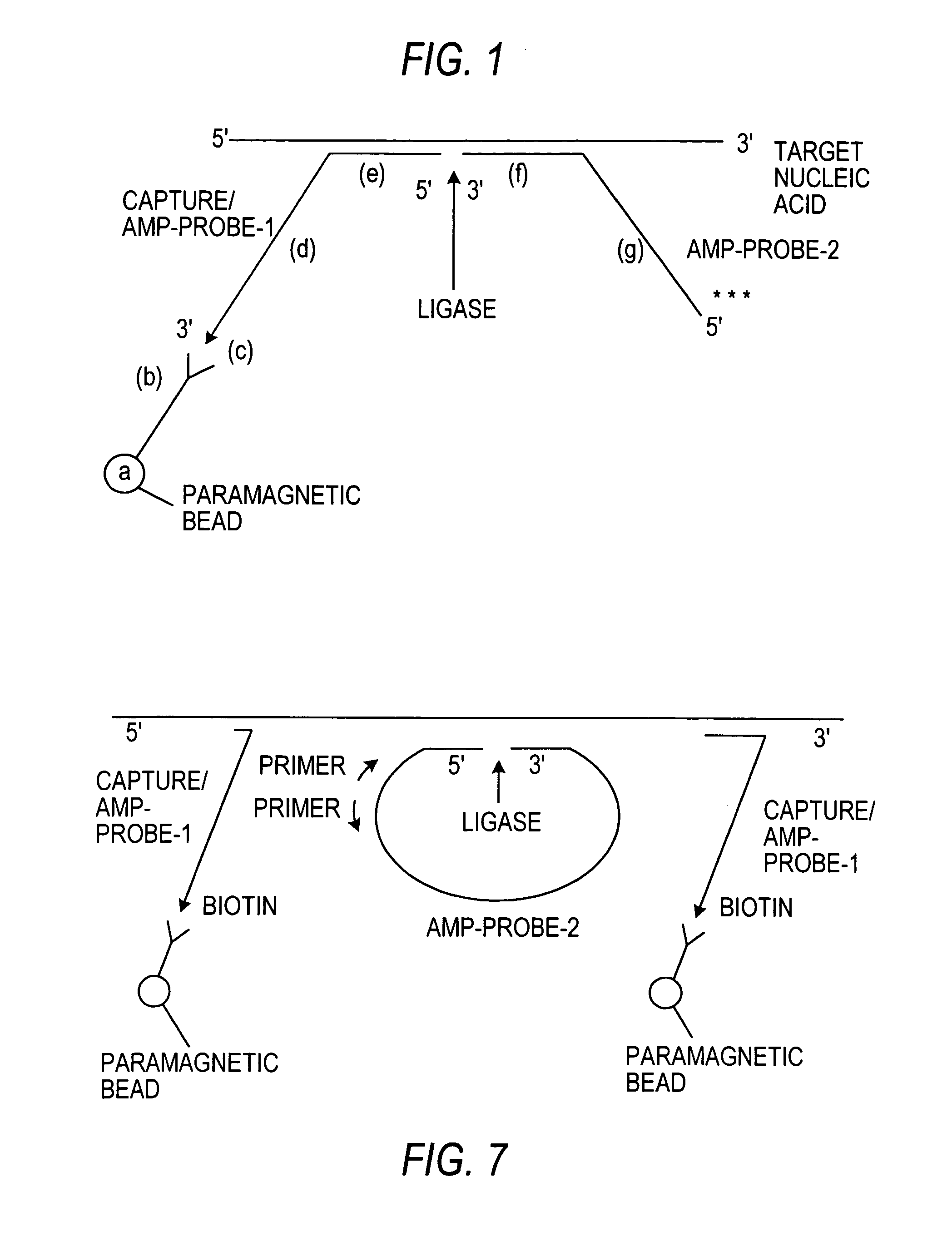
[0182]A pair of oligodeoxyribonucleotide probes, designated Capture / Amp-probe-1 (HIV) and Amp-probe-2 (HIV), respectively for detecting the gag region of HIV-1 RNA were prepared by automated synthesis via an automated DNA synthesizer (Applied Biosystems, Inc.) using known oligonucleotide synthetic techniques. Capture / Amp-probe-1 (HIV) is an oligodeoxyribonucleotide comprising 59 nucleotides and a 3′ biotin moiety, which is added by using a 3′-biotinylated nucleoside triphosphate as the last step in the synthesis. The Capture / Amp-probe-1 (HIV) used in this Example has the following nucleotide sequence (also listed below as SEQ ID NO. 1):
1 11 215′- CCATCTTCCT GCTAATTTTA AGACCTGGTA 31 41 51 ACAGGATTTC CCCGGGAATT CAAGCTTGG -3′
[0183]The nucleotides at positions 24-59 comprise the generic 3′ end of the probe. Within this region are recognition sequences for SmaI (CCCGGG), EcoR...
example 2
Direct Detection of HIV-1 RNA in a Sample
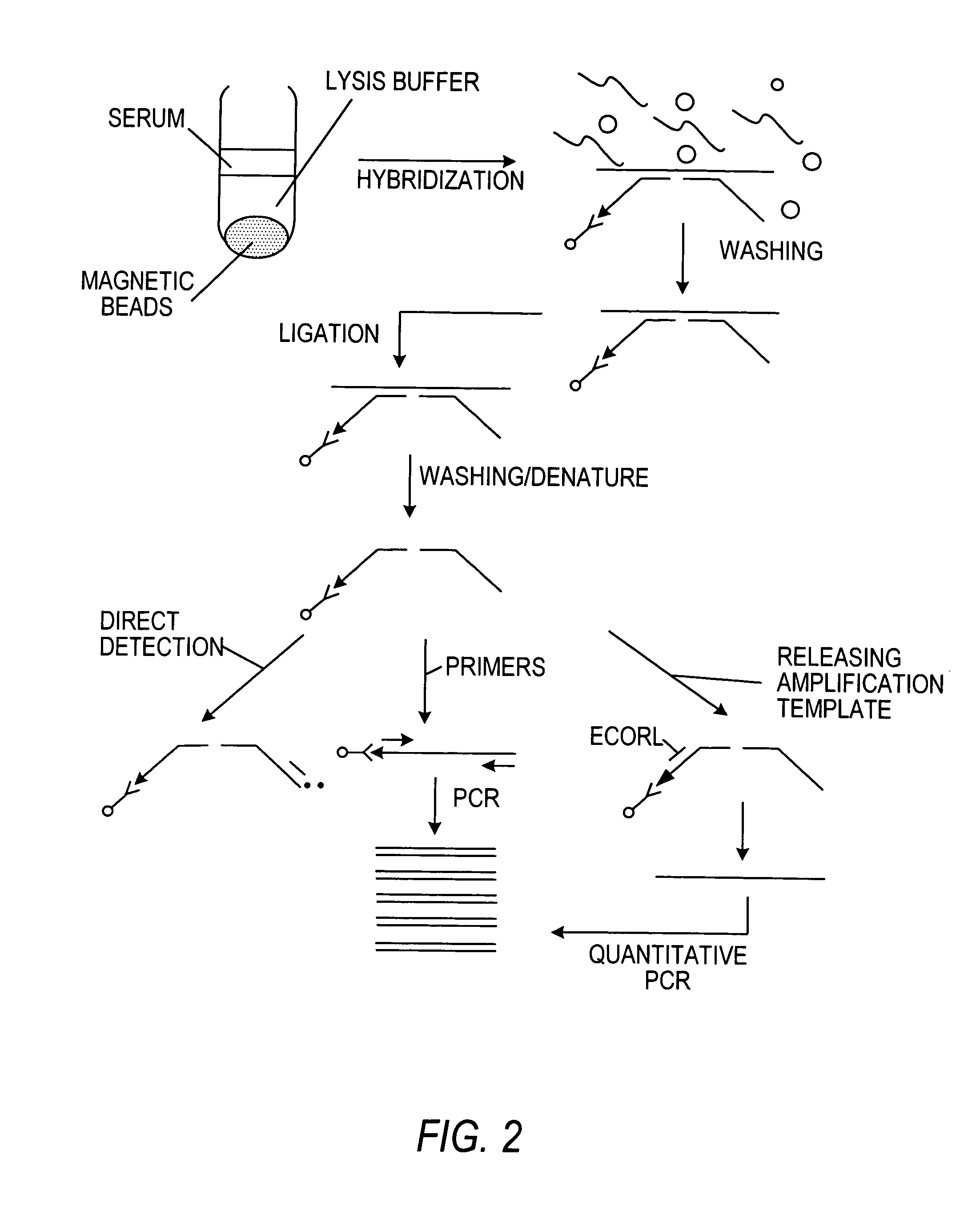
[0197]The ability of the present method to directly detect the presence of HIV-1 RNA in a sample was also determined. The probes used in this Example are the same as in Example 1 (SEQ ID NOS. 1 and 2). For direct detection, Amp-probe-2 (HIV) (SEQ ID NO. 2) was labeled at its 5′ end with 32P by the T4 phosphokinase reaction using 32P-γ-ATP. The various reaction mixtures were as follows:
[0198]Streptavidin-coated paramagnetic beads, 3′-biotinylated Capture / Amp-probe-1 (HIV) (SEQ ID NO. 1), Amp-probe-2 (HIV) (SEQ ID NO. 2) 5′(32P), HIV-1 RNA transcript.
[0199]Streptavidin-coated paramagnetic beads, 3′-biotinylated Capture / Amp-probe-1 (HIV), Amp-probe-2 (HIV) 5′(32P).
[0200]Streptavidin-coated paramagnetic beads, Amp-probe-2 (HIV) 5′(32P), HIV-1 RNA transcript.
[0201]Hybridizations, using each of the above three reaction mixtures, were carried out in 200 of a 1M GnSCN buffer comprising 1M GnSCN, 0.5% NP-40 (Nonidet P-40, N-lauroylsarcosine, Sigma Che...
example 3
Detection of Mycobacterium
Avium-Intracellulaire (MAI) in Patient Samples
[0203]A recent paper (Boddinghaus et al., J. Clin. Microbiol. 28:1751, 1990) has reported successful identification of Mycobacteria species and differentiation among the species by amplification of 16S ribosomal RNAs (rRNAs). The advantages of using bacterial 16S rRNAs as targets for amplification and identification were provided by Rogall et al., J. Gen. Microbiol., 136:1915, 1990 as follows: 1) rRNA is an essential constituent of bacterial ribosomes; 2) comparative analysis of rRNA sequences reveals some stretches of highly conserved sequences and other stretches having a considerable amount of variability; 3) rRNA is present in large copy numbers, i.e. 103 to 104 molecules per cell, thus facilitating the development of sensitive detection assays; 4) the nucleotide sequence of 16S rRNA can be rapidly determined without any cloning procedures and the sequence of most Mycobacterial 16S rRNAs are known.
[0204]Thre...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com