Surface concatemerization of templates
a surface concatenation and template technology, applied in the field of surface concatenation of templates, can solve the problems of limiting read length, increasing the burden on the robustness of sequencing biochemistry, and daunting task of cataloguing human genetic variation and correlating this variation with susceptibility to disease, etc., to achieve the effect of improving densities, improving read quality, and prolonging read length
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
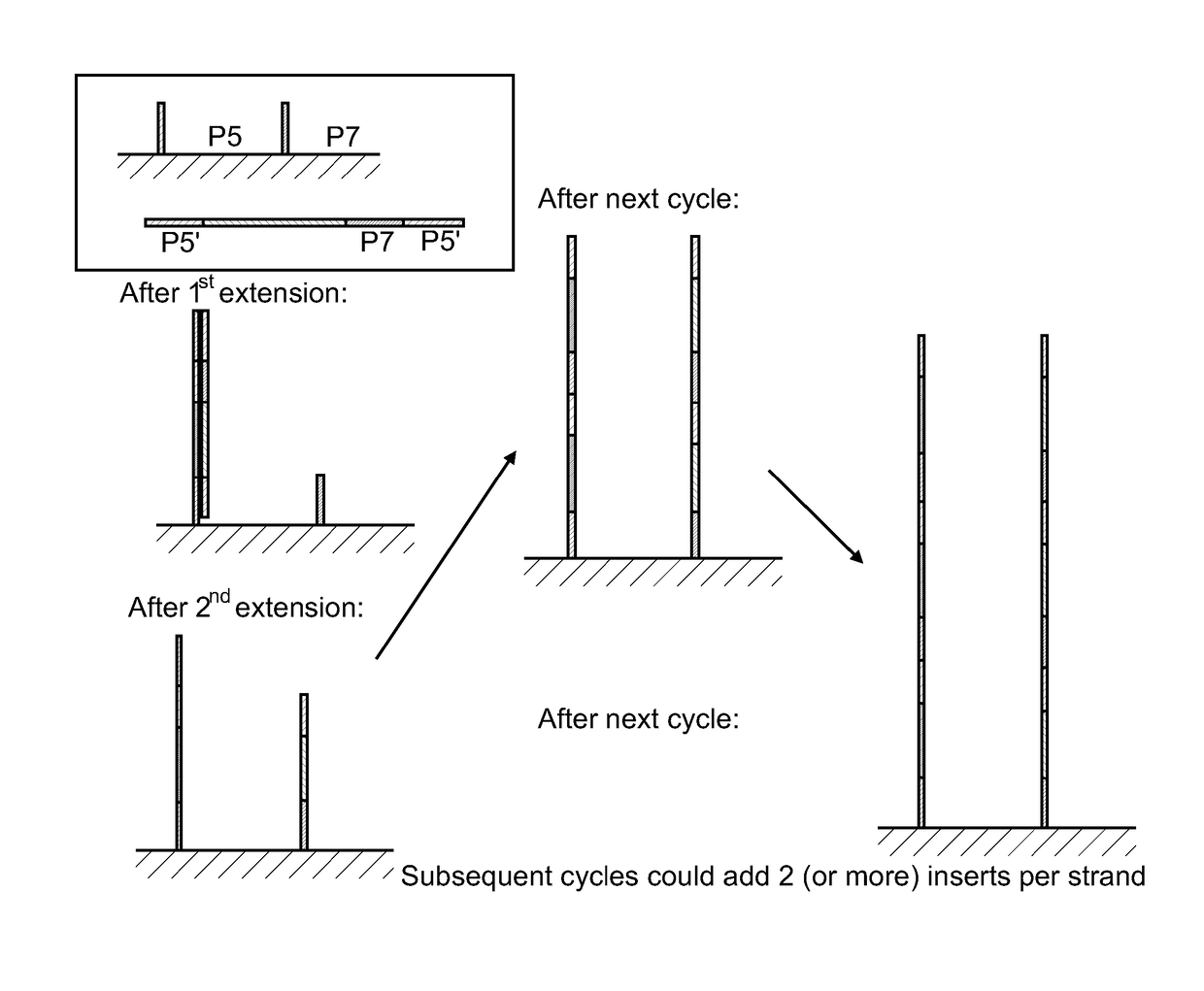
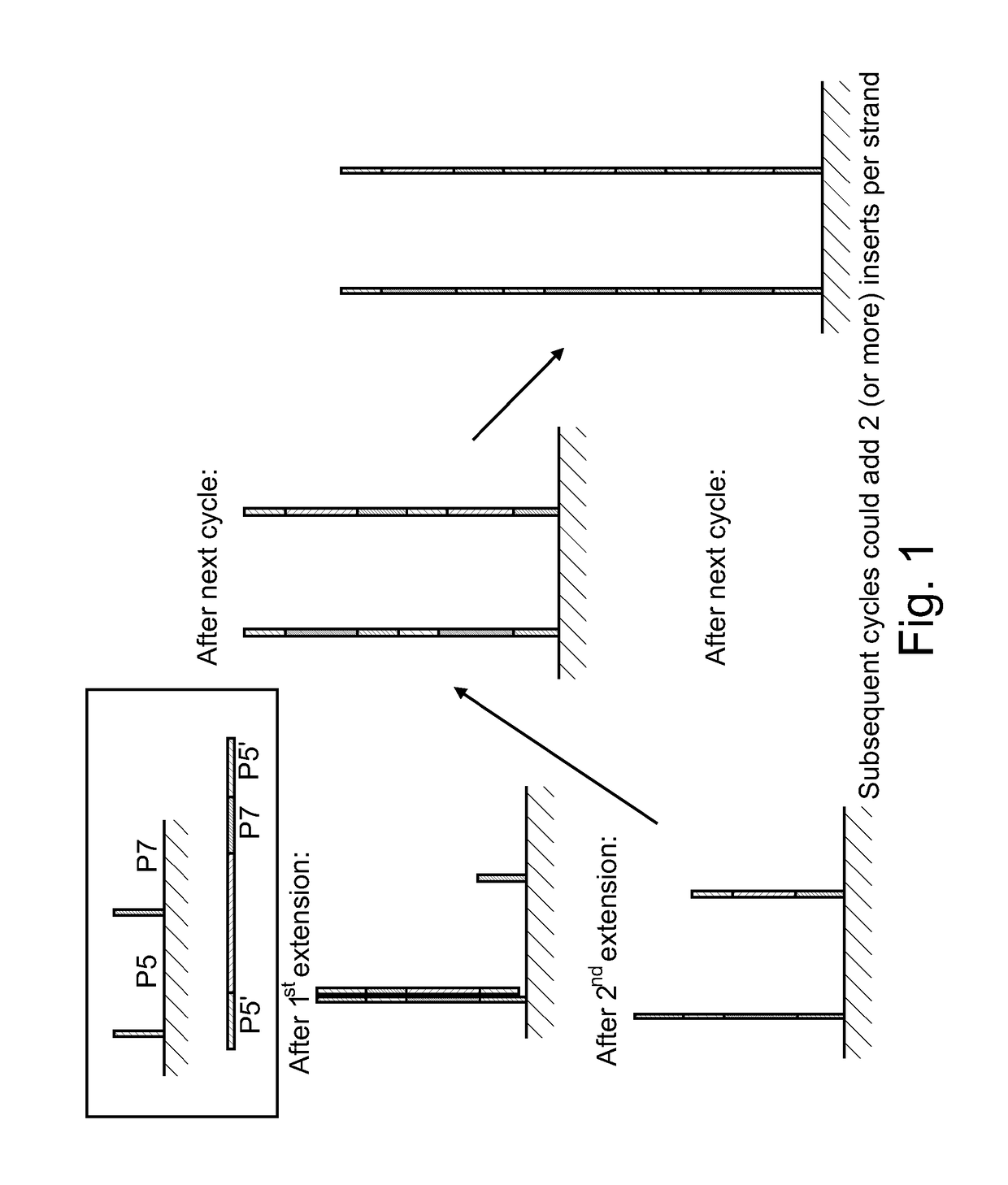
[0075]This example describes solid phase amplification according to one embodiment, as illustrated in FIG. 1. A standard universal PhiX or CT418 library containing P5 and P7′ regions was used to attach the additional P5′ at the 5′ end of the already prepared library. This was accomplished by performing additional 18 PCR cycles. The libraries were diluted to 200 pM final and amplified using either standard primer or P7-P5′ primer. Each 50 ul PCR reaction contained 22 μl of H2O, 25 μl of 2× Phusion Mastermix (NEB), 1 μl each of the appropriate primer and DNA. After PCR, the resulting library concentrations were determined by the Nanodrop (Thermo Scientific) and diluted to 10 nM in buffer EB (QIAGEN)+0.05% Tween20. A flowcell was prepared by grafting HEG primers (lanes 1-4) or standard PE primers (lanes 5-8) using the protocol described in U.S. Pat. Nos. 8,536,477, 8,715,966, and U.S. Patent Application Pub. 2008 / 0280773, the content of each of which is incorporated by reference herein...
example 2
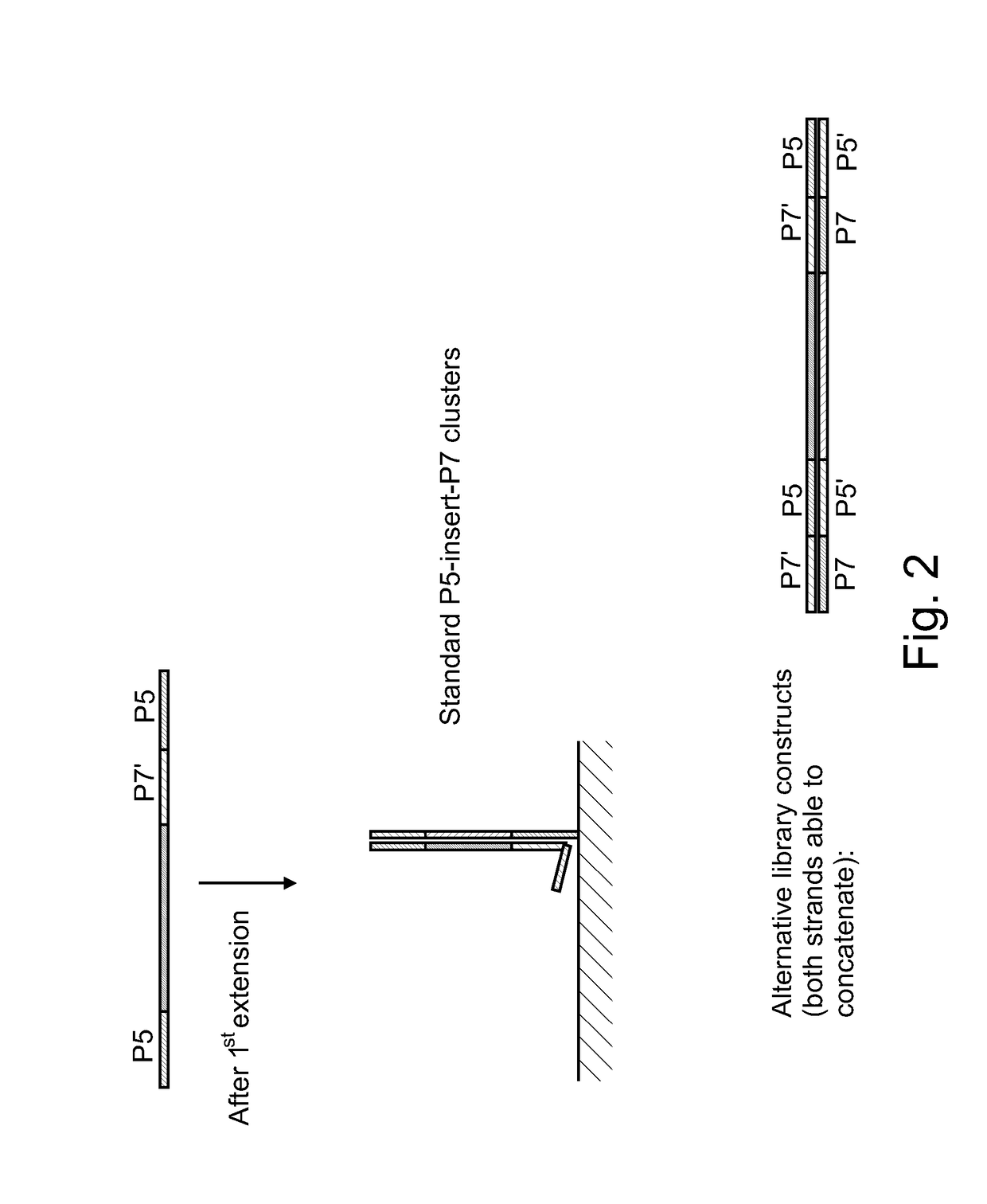
[0078]This example describes solid phase amplification according to an embodiment, as illustrated in FIG. 3. The experiments reported in FIG. 6 show the results from concatemerization of 20T / A linkers on the P5 / P7 grafting primers (referred as 20T-P7 / 20A-P5) instead of attaching the concatamer facilitating primers on to the library during PCR process. In this experiment, the flowcell was grafted with 20A / T Paired-end P5 / P7 primers alongside standard primers. The standard grafting protocol as described in incorporated materials of U.S. Pat. Nos. 8,536,477, 8,715,966, and U.S. Patent Application Pub. 2008 / 0280773 was followed and this grafted flowcell was used for cluster amplification on a cBOT clonal amplification system (Illumina Cat # SY-301-2002) using the TruSeq cluster generation kit for Genome Analyzer (Illumina) as per manufacturer's recommended protocol. CT418 libraries were used as template; amplified 105 cycles at 60° C. The flowcell was stained with SYBR Green and imaged ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| sizes | aaaaa | aaaaa |
| sizes | aaaaa | aaaaa |
| sizes | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com