Early post-transfection isolation of cells (EPIC) for biologics production
a cell and post-transfection technology, applied in the field of early post-transfection isolation of cells (epic) for biologics production, can solve the problems of drug-based selection time-consuming, negative impact on clonal stability, and inability to meet the needs of targeted populations, and achieve the effect of improving the production of target polypeptides
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Sorting for Early Post-Transfection Isolation of Cells (EPIC)—Proof of Concept
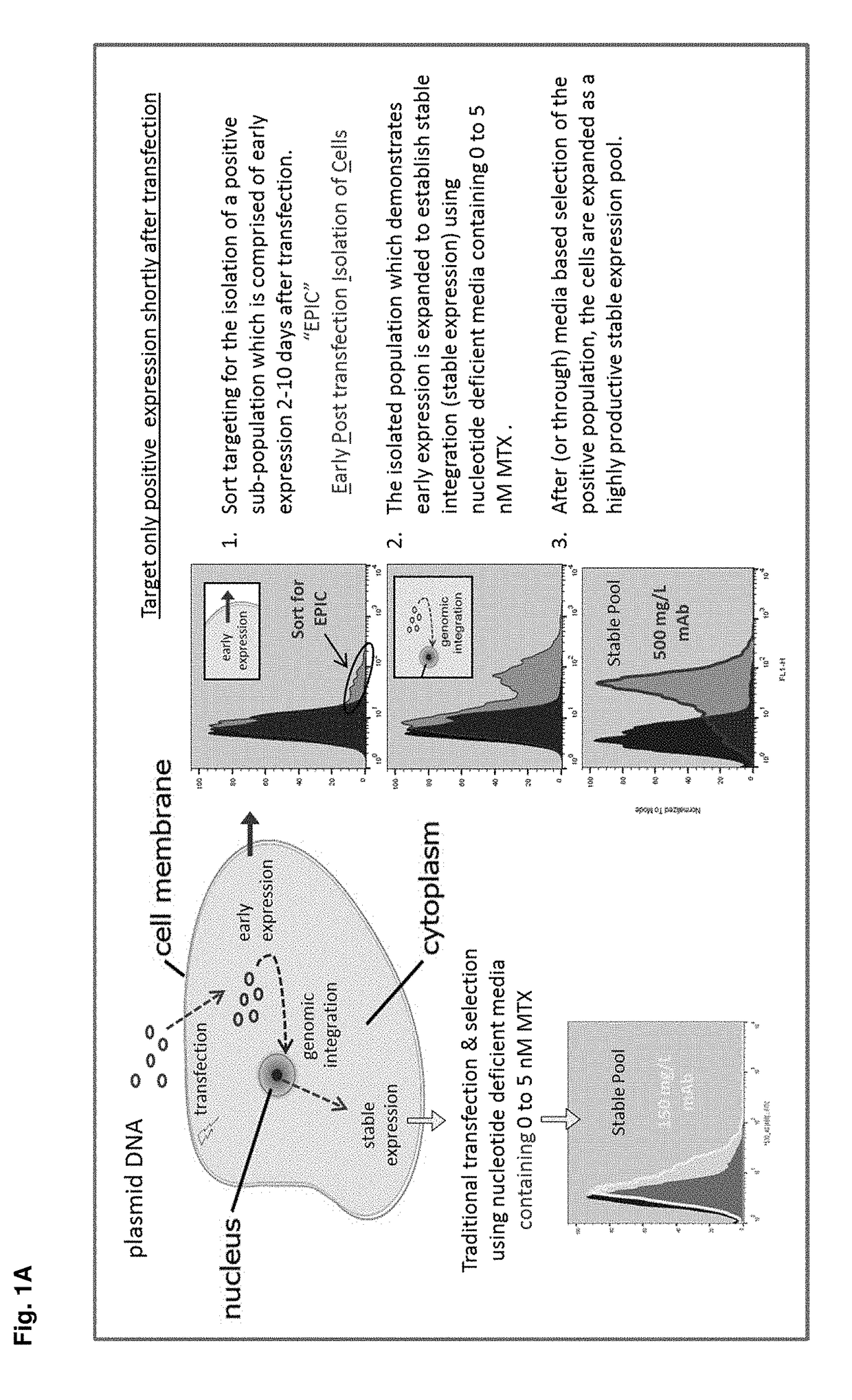
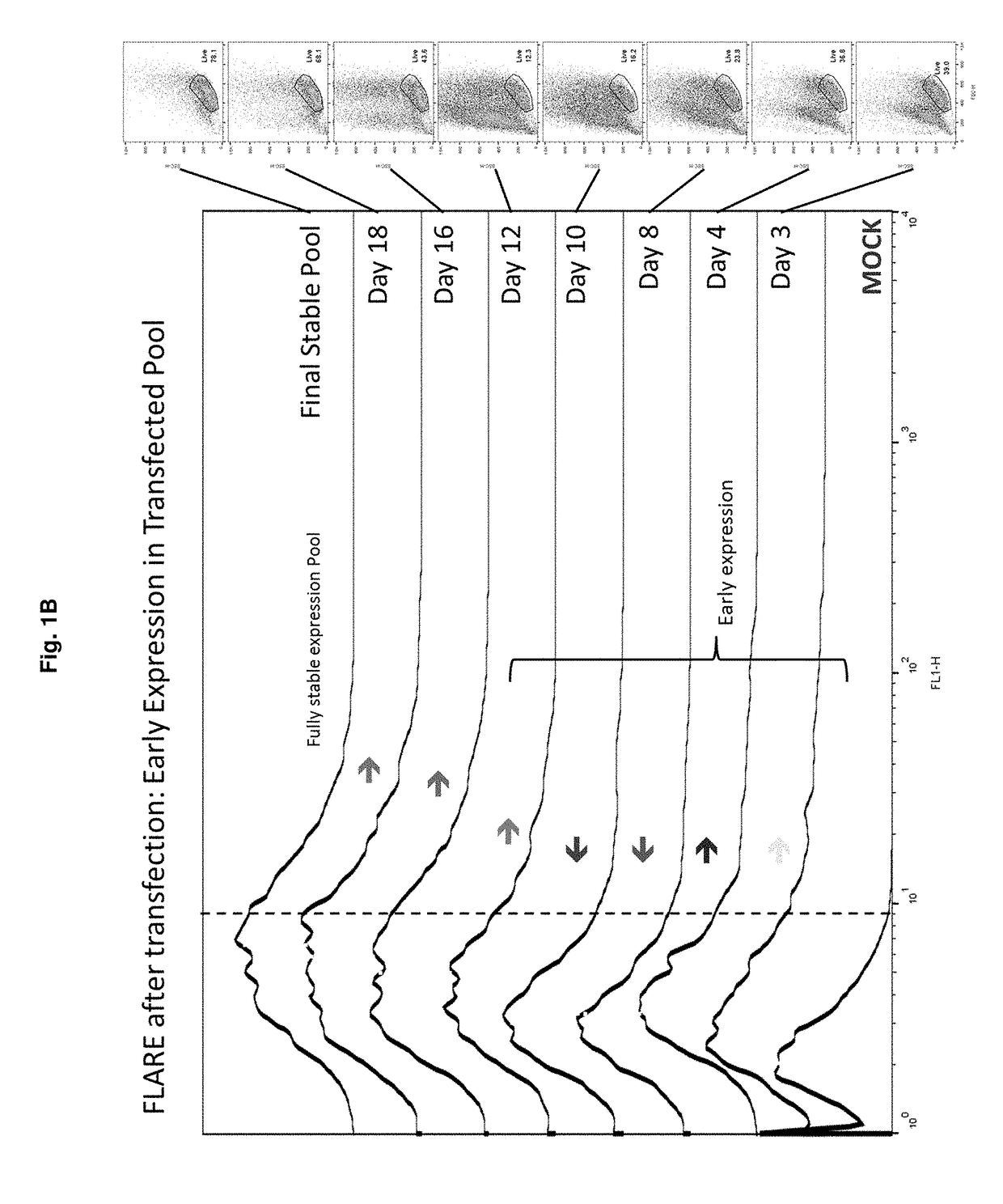
[0143]This example demonstrates the feasibility of a method of sorting to target an unselected transfected early-expressing population for bulk enrichment prior to selection. This method of sorting is called “short sorting” or “EPIC” (Early Post-transfection Isolation of Cells) and is designed to sort-isolate, or bulk enrich, early reporter expression shortly after transfection. EPIC may significantly reduce selection timelines and / or improve productivity of the resulting heterogeneous population. Experiments have been performed to investigate the reporter expression profile of a transfected population throughout the course of a nucleotide-deficient selection process. FIG. 1A depicts a general scheme for EPIC. FIG. 1B shows the early expression of the reporter gene during the nucleotide-deficient selection process. These offset histograms demonstrate that early expression (e.g. day 3-4) is positive and sor...
example 2
Sorting for Early Post-Transfection Isolation of Cells (EPIC)—Producer Cell Pool Generation
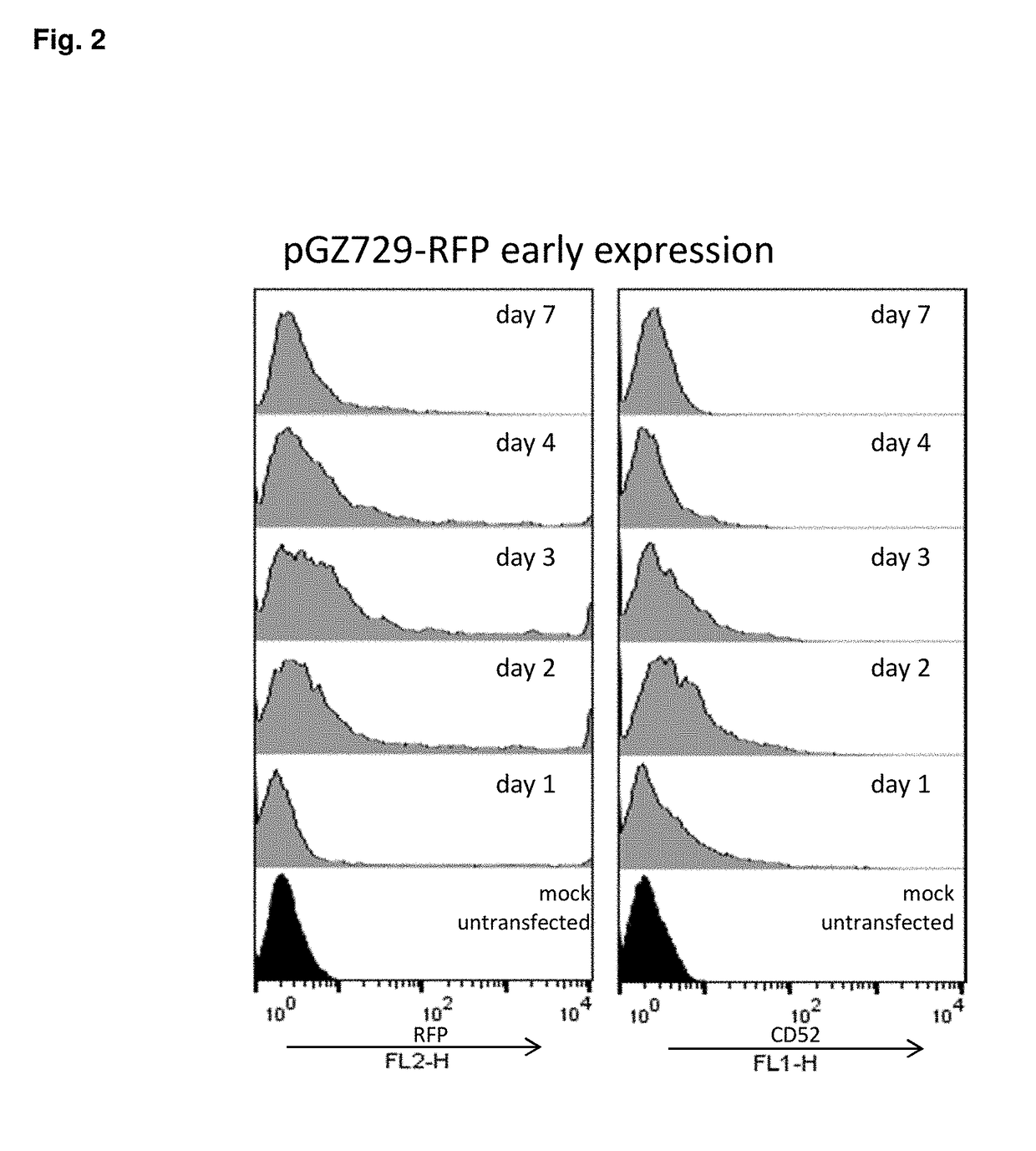
[0159]EPIC was initially attempted using mAb#1 in which CHO cells were transfected and given 2 days to recover, after which 0 nM MTX selection was initiated to establish early expression. Four days after transfection, early expression of CD52 cell surface reporter was targeted for sort isolation (EPIC). Sorting targeted only positive expression which was collected as a bulk enriched population of about 1 million cells which was then allowed to continue selection in nucleotide-deficient media (0 nM MTX). As a control, a non-sorted transfection was allowed to continue selection via standard selection procedure. As shown in FIG. 4, by day 8 sorting for EPIC yielded a slightly enriched population as seen by CD52 reporter expression as compared to standard selection. As selection of both populations continued, however, this small EPIC sub-population became more prominent over time. In fact, the CD5...
example 3
Sorting for Early Post-Transfection Isolation of Cells (EPIC)—Clone Generation
[0162]The EPIC-generated pool of Example 2 was next used to generate clones using FLARE as previously described (see, e.g., Cairns, V. et al. (2011) Utilization of Non-AUG Initiation Codons in a Flow Cytometric Method for Efficient Selection of Recombinant Cell Lines, Biotechnol Bioeng 108(11):2611-2622). Briefly, FLARE was used to isolate and single cell plate the top 3-5% of reporter-expressing cells from each pool using FACS. Expanded clones were then screened (taking top 30% positive expressers), again using FLARE, to identify only the top tier clones to expand for target polypeptide titer evaluation. As shown in FIG. 6, top expressing EPIC-generated clones achieved similar titers to those of best clones from traditional methods, e.g., using MTX-amplified pools (near 2.0 g / L). Results demonstrated that using EPIC to isolate early expression populations prior to selection is a viable alternative to trad...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Time | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com