Hydroxymethylation analysis of cell-free nucleic acid samples for assigning tissue of origin, and related methods of use
a cell-free nucleic acid and hydroxymethylation technology, applied in the field of biotechnology, can solve the problems of unique challenges in the analysis of cell-free dna, and achieve the effects of improving the quality of cell-free dna
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
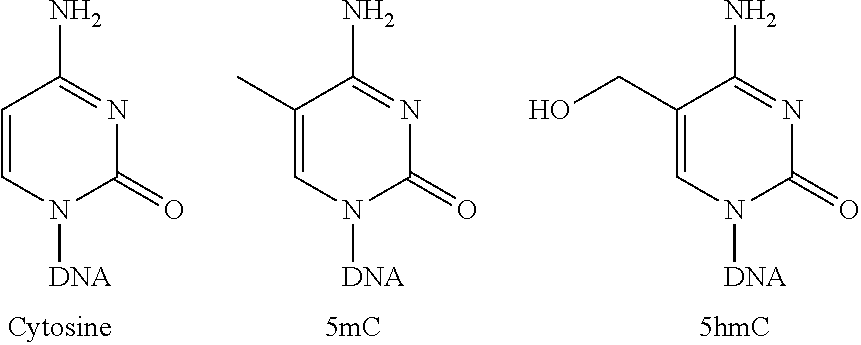
[0069]The invention provides a method for probabilistically assigning a tissue of origin to a nucleic acid in a cell-free fluid sample, e.g., a cell-free DNA sample obtained from a human patient. Related methods and systems are also provided, involving diagnosis, prognosis, patient monitoring, and other methods. The invention assesses the similarity of a detected hydroxymethylation profile of a nucleic acid across a large reference set of hydroxymethylation profiles, i.e., a hydroxymethylome data set containing hydroxymethylation data for a plurality of loci on each of a plurality of tissue-specific genes, for a plurality of tissue types.
I. Terminology:
[0070]Unless defined otherwise, all technical and scientific terms used herein have the meaning commonly understood by one of ordinary skill in the art to which the invention pertains. Specific terminology of particular importance to the description of the present invention is defined below. Other relevant terminology is defined in In...
PUM
| Property | Measurement | Unit |
|---|---|---|
| hydroxymethylation density | aaaaa | aaaaa |
| density | aaaaa | aaaaa |
| nucleic acid length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com