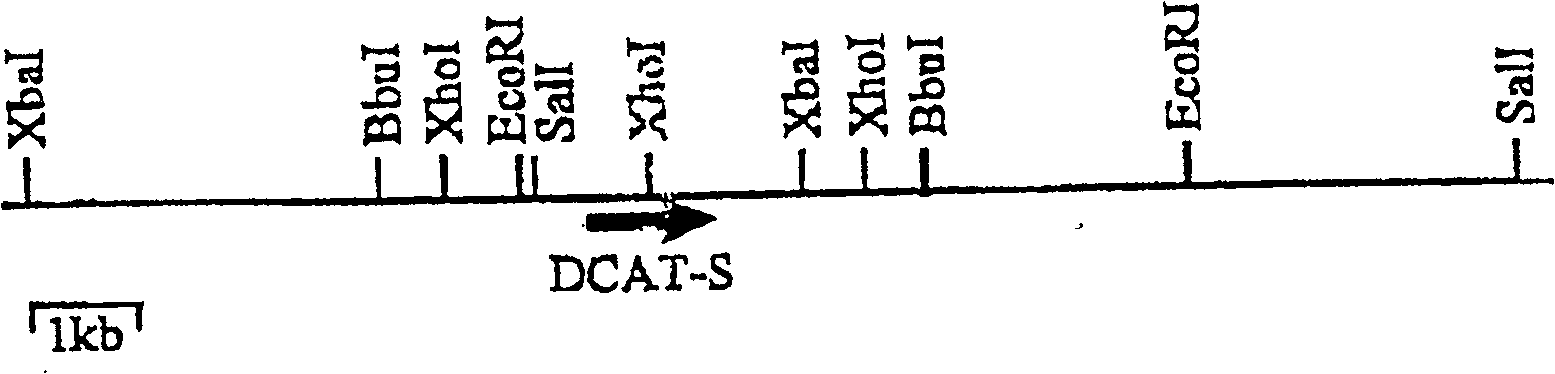
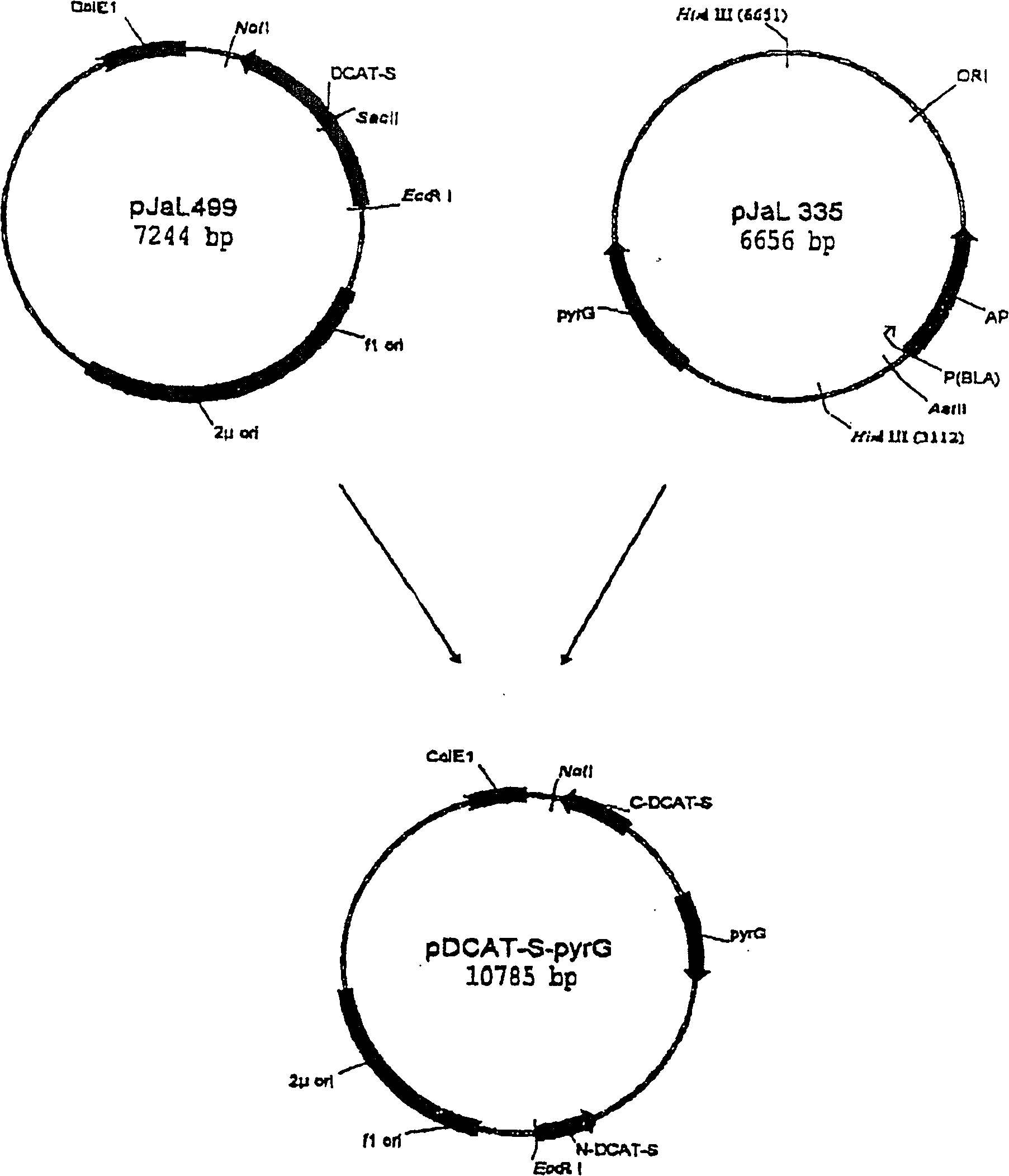
Methods for producing polypeptides in aspergillus mutant cells
A technology of Aspergillus and cells, applied in the direction of microorganism-based methods, methods using fungi, biochemical equipment and methods, etc., can solve problems that affect the cost of products and the time to put products on the market
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0236] A. Construction of a CPA-negative strain of Aspergillus oryzae Bz 14
[0237] Freeze-dried spores of Aspergillus oryzae Bz 14 strain were irradiated with an optimal dose of γ-rays, ranging from 1000Gy to 1250Gy, and these spores were plated on the plates of screening medium 1 at a density of 25-50 colonies / 9cm plate superior. Cyclopiazonic acid producing colonies formed a red counter (bottom of the colony) on selection medium 1 due to the presence of red insoluble CPA-Fe complexes.
[0238] Approximately 50,000 colonies were screened from irradiated spores and 154 CPA-deficient colonies were isolated, which were characterized by a cream / white appearance. After re-isolation, 64 strains still maintained a non-red appearance on the selection medium 1 plates. CPA was not detected in 52 strains using TLC-plug (plate) analysis. These strains were then cultured in MDU-1B medium under toxin-induced (34° C., 250 rpm, cultured for 5 days) shaking flask fermentation culture con...
Embodiment 2
[0255] Expression of lipase in Aspergillus oryzae strains JAL228 and BECh1
[0256] a. Construction of plasmid pCaHj493
[0257] The lipase plasmid pAHL (WO 97 / 07202) was digested with BamHI and SalI, and a 916bp lipase-encoding fragment was isolated.
[0258] pCaHj 483 was digested with BamHI and XhoI and the 6757 bp vector fragment was ligated to the lipase fragment as described in WO 98 / 00529. The ligation mixture was used to transform E. coli DH5α cells, and transformants containing the desired plasmid were isolated. This plasmid was named pCaHj493.
[0259] b. Transformation of pCaHj 493 into JaL228 and BECh-1 strains
[0260] pCaHj 493 was transformed into Aspergillus oryzae strains JaL228 and BECh1 by selection in acetamide as described in EP-A-0531372. Transformants were re-spored twice. Second reisolated spores of each transformant were tested for lipase production in shake flask and microtiter plate cultures.
Embodiment 3
[0262] A. Production of lipase in CPA-negative and CPA-positive Aspergillus oryzae strains
[0263] The lipase production of 18 JaL228 transformants and 30 BECh 1 transformants prepared as described in Example 2 in shake flask cultures was examined.
[0264] Cove N slant cultures of transformants were harvested with 10 ml of 0.1% Tween solution, and the spore suspension was used as inoculum in 100 ml of Gl-Gly medium and placed in 500 ml double baffle shake flasks. The medium was incubated on a rotary shaker at 250 rpm at 34°C for 24 hours. Then 10ml of the Gl-Gly culture was transferred to 100ml of 1 / 5MDU-2BP in a 500ml shake flask and continued at 34°C at 250rpm.
[0265] Samples were removed after 50 hours, filtered through Miracloth and centrifuged at 4000 xg. With a simple radioimmunodiffusion method (see Scand.J.Immuno.Vol.17, suppl.10, pages 41-56, (1983) "Technical Handbook of Immunoprecipitation Gel Technology", edited by N.H.Axelsen, Blackwell Scientific Publicatio...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap