Constructing 9LLUC cell strain of expressing luciferase stably, and application
A luciferase and stable expression technology, applied in the field of biomedicine, can solve the problems such as the inability to obtain stable expression, the inability to complete the preparation and real-time observation of glioma animal models, and the inability to establish stable expression cell lines, and achieves a promising application prospect. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
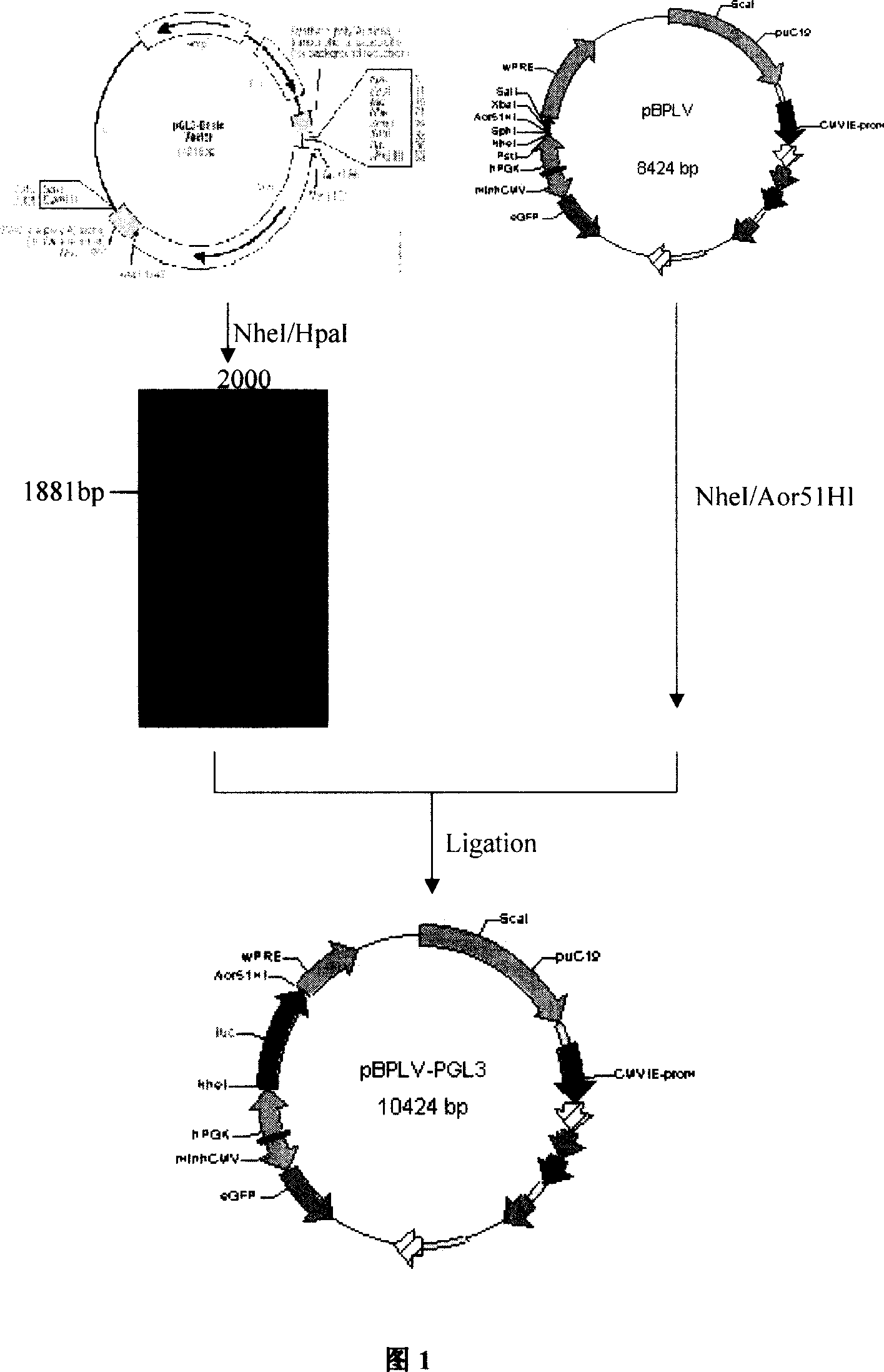
[0019] Example 1. Construction of lentiviral expression plasmid pBPLV-LUC.
[0020] The lentiviral expression vector pBPLV was constructed according to the method introduced by Mario Amendola et al. (Nature Biotechnology, 2005, 23: 108-116). After digesting the MA1 expression plasmid with PstI restriction enzyme and SalI restriction endonuclease, the multi-cloning site sequence (5'-GGCTAGCATGCTCTAGAGCGCTG-3', 3'-ACGTCCGATCGTACGAGATCTCGCGACAGCT-5') was added to construct the lentiviral expression plasmid pBPLV.
[0021] Firefly luciferase PGL 3 -Basic plasmid was purchased from Promega Company, passed NheI restriction endonuclease (Takara Company, D1162A), HpaI restriction endonuclease (Takara Company, D1064A) each 2 μl, PGL 3 -Basic plasmid 30 μl, 10×T Buffer 5 μl, incubate at 37°C for 2 hours to recover the LUC gene fragment, the fragment size is 1881bp; lentiviral expression vector pBPLV 30 μl, NheI restriction endonuclease 2 μl, Aor51 HI restriction endonuclease ( Takara...
Embodiment 2、293
[0022] The packaging of embodiment 2, 293FT cells
[0023] 293FT cells were cultured in high glucose DMEM containing 10% fetal bovine serum (FBS). In a sterile 5ml tube, add 1.5ml of serum-free DMEM medium, and then add the packaging plasmid mixture 4.2μg pLP1, 2μgpLP2, 2.8μg pLP / VSVG (Virapower TM Lentiviral Packaging Mix, Invitrogen Company, 4975-00) and 5 μg lentiviral expression plasmid, mixed gently. In another sterile 5ml tube, dilute 42μl Lipofectamine 2000 (Invitrogen, 11668-027) in 1.5ml serum-free DMEM medium, mix gently, and place at room temperature for 5min. Mix the above 2 tubes of liquid and mix gently. Incubate at room temperature for 20 min to form DNA-Lipofectamine 2000 complex. During DNA-lipid complex formation, 293FT cells were digested with 0.25% trypsin, counted, and resuspended in high-glucose DMEM containing 10% serum to a density of 1.2×10 6 pieces / ml. Add the DNA-Lipofectamine 2000 complex to a 10cm cell culture dish containing 5ml of growth me...
Embodiment 3
[0024] Example 3, 9L LUC Establishment of cell lines.
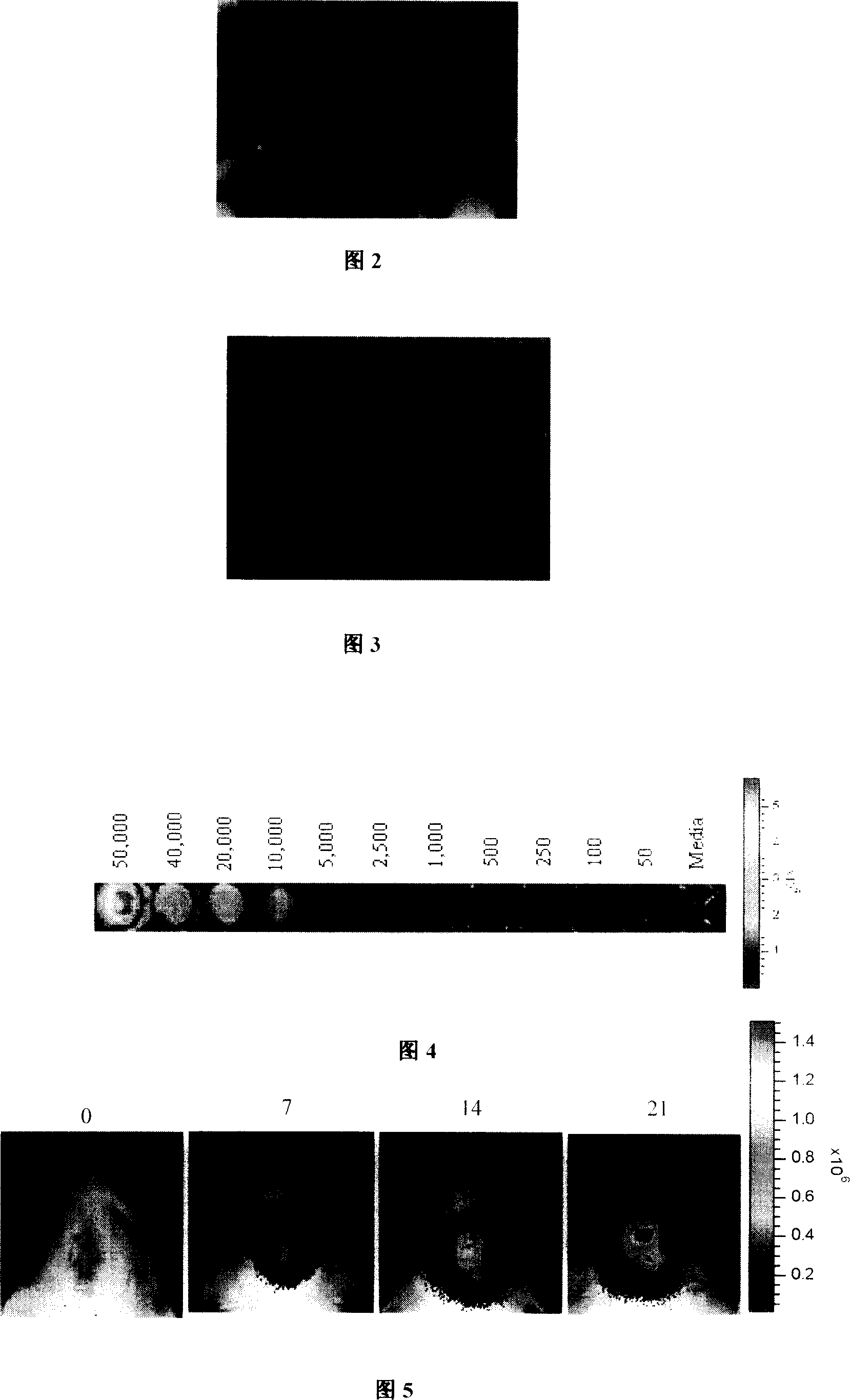
[0025] 9L cells in good growth state were routinely cultured for 24 hours and then digested with 0.25% trypsin. They were added to the culture dish together with 1ml of the prepared lentivirus solution and Polybrene (final concentration: 8 μg / ml), and were routinely cultured overnight. Fresh growth medium was replaced every The medium was changed once every 3 days. After 14 days of culture, the cells were digested to prepare a single cell suspension, and the 9L cells with green fluorescent protein were sorted by flow cytometry. LUC After the cells were cultured to 80-90% confluence, they were subcultured at a ratio of 1:2 (results shown in Figure 3).
[0026] Example 4, 9L LUC detection of luciferase. Will 9L LUC The cells were serially diluted in a 96-well plate, respectively 50000, 40000, 20000, 10000, 5000, 2000, 1000, 500, 200, 100, 50, media / well, and the firefly luciferase substrate D-luciferin (150 μg / ml, Sig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com