Fast high-flux gene site-directed mutagenesis method
A high-throughput, genetic technology, applied in the field of genetic manipulation, can solve problems such as the impact of PCR amplification efficiency and mutagenesis efficiency, difficulty in gene insertion and deletion mutations, and difficult amplification of plasmid vectors, so as to save digestion steps, The effect of improving mutagenesis efficiency and shortening the experimental period
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
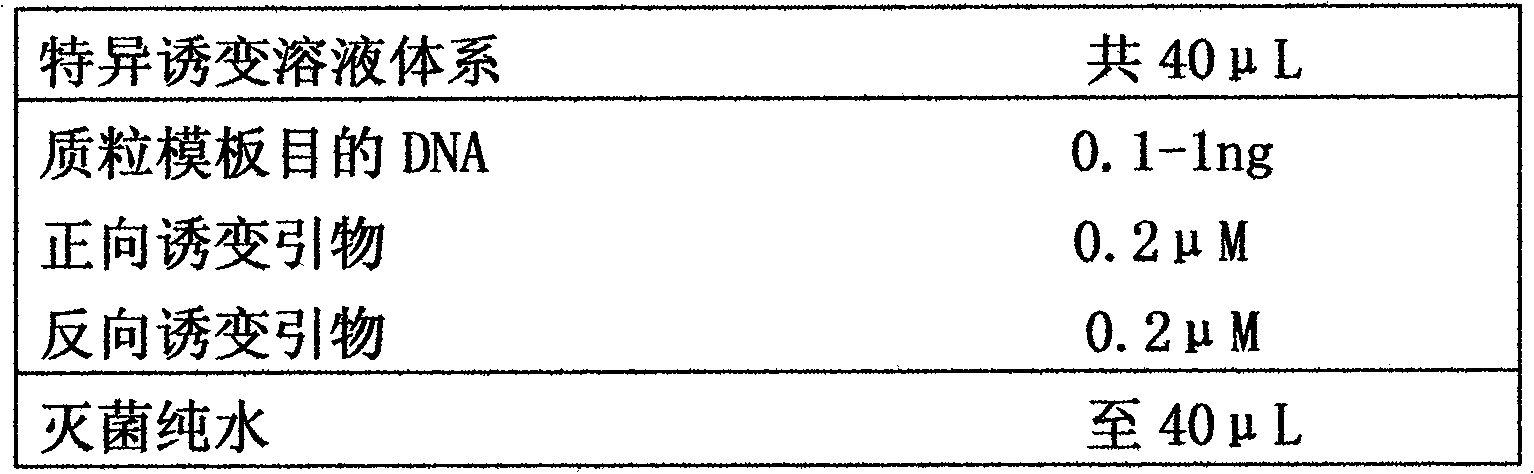
[0031] The gene thyA cloned on pBluescript SKII is deleted and mutated by using the method of the present invention, and a mutant plasmid with 726 nucleotides missing is successfully obtained.
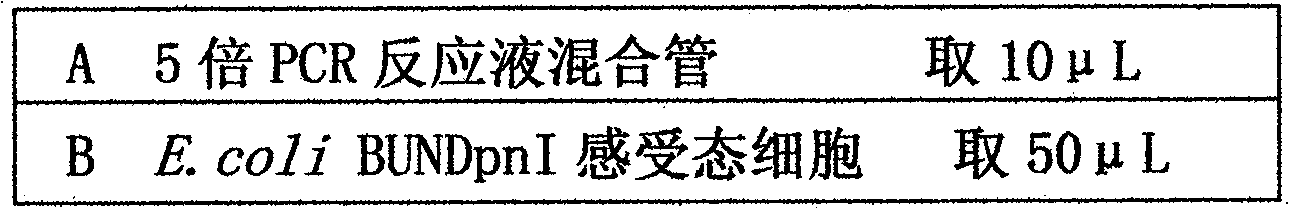
[0032] 1. Construct the strain E.coli BUNDpnI expressing DNA adenine methylase deficiency of DpnI.
[0033] Design a pair of primers at both ends of the kanamycin expression unit, with a 50nt homologous region with the gene encoding DNA adenine methylase at both ends of the primers, use the plasmid pShuttle as a template, and carry out with this pair of primers The amplified PCR fragment is mediated by λ-Red recombinase to replace the gene encoding DNA adenine methylase on the E.coli BUN21 chromosome with the kanamycin gene, and the DNA adenine methylase deficiency is first obtained strains. Secondly, the PCR method is used to synthesize the gene dpnc encoding DpnI, and this gene is cloned into the temperature-sensitive plasmid pIJ790 (the replication origin of this plasmid is a tempe...
Embodiment 2
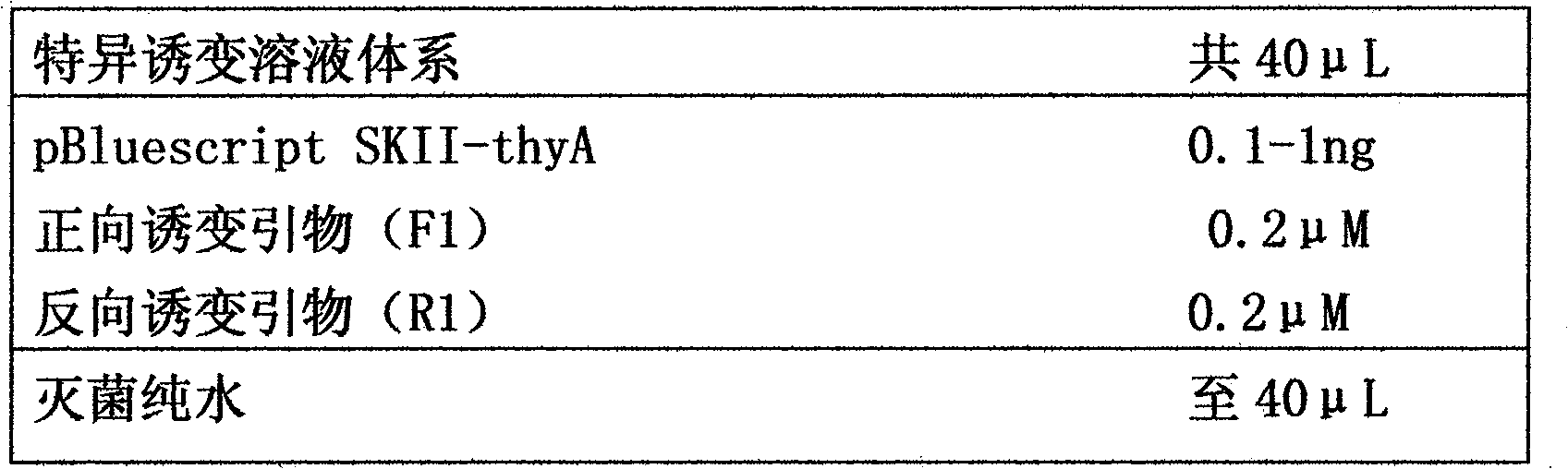
[0063] Using this method, a PvuII restriction site in pBluescript SKII-thyA was point-mutated, and a plasmid with the mutation of the restriction site was successfully obtained.
[0064] 1. Using the DpnI-expressing DNA adenine methylase-deficient strain E.coli BUNDpnI constructed in Example 1.
[0065] 2. Design partially overlapping (15nt) mutagenic primers.
[0066] Analyze the DNA sequence of pBluescript SKII-thyA, and design a pair of primers (forward mutagenesis primer F2 and reverse mutagenesis primer R2) at the site where the point mutation will be carried out. Both F2 and R2 can match the mutation site. The nucleotide sequence at the site to be mutated is also included in the pair of primers in complementary form (indicated in lowercase letters), F2 and R2 overlap 15nt at the 5' end (indicated in bold font) .
[0067] The primer sequences were designed as:
[0068] The forward primer is: 5-' AAACGACCC-3',
[0069] The reverse primer is: 5-' ACCGTAGTGA-3'
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com