Co-modified nucleic acid segment of locked nucleic acid and minor groove conjugation
A technology of nucleic acid fragments and co-modification, which is applied in the direction of DNA/RNA fragments, sugar derivatives, biochemical equipment and methods, etc. It can solve the problems of limited annealing temperature and inability to fully meet the design requirements, and achieve the effect of enhanced recognition ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
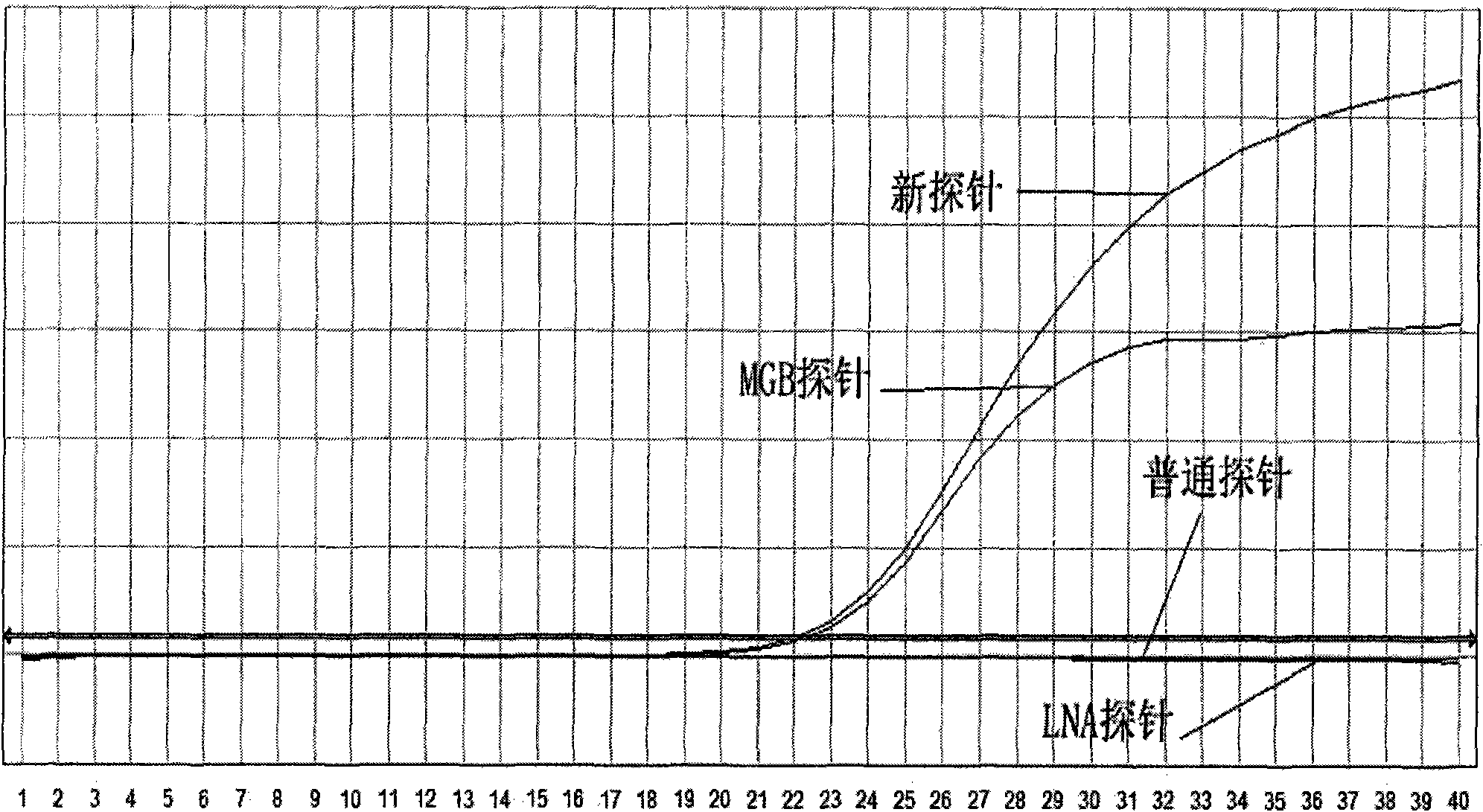
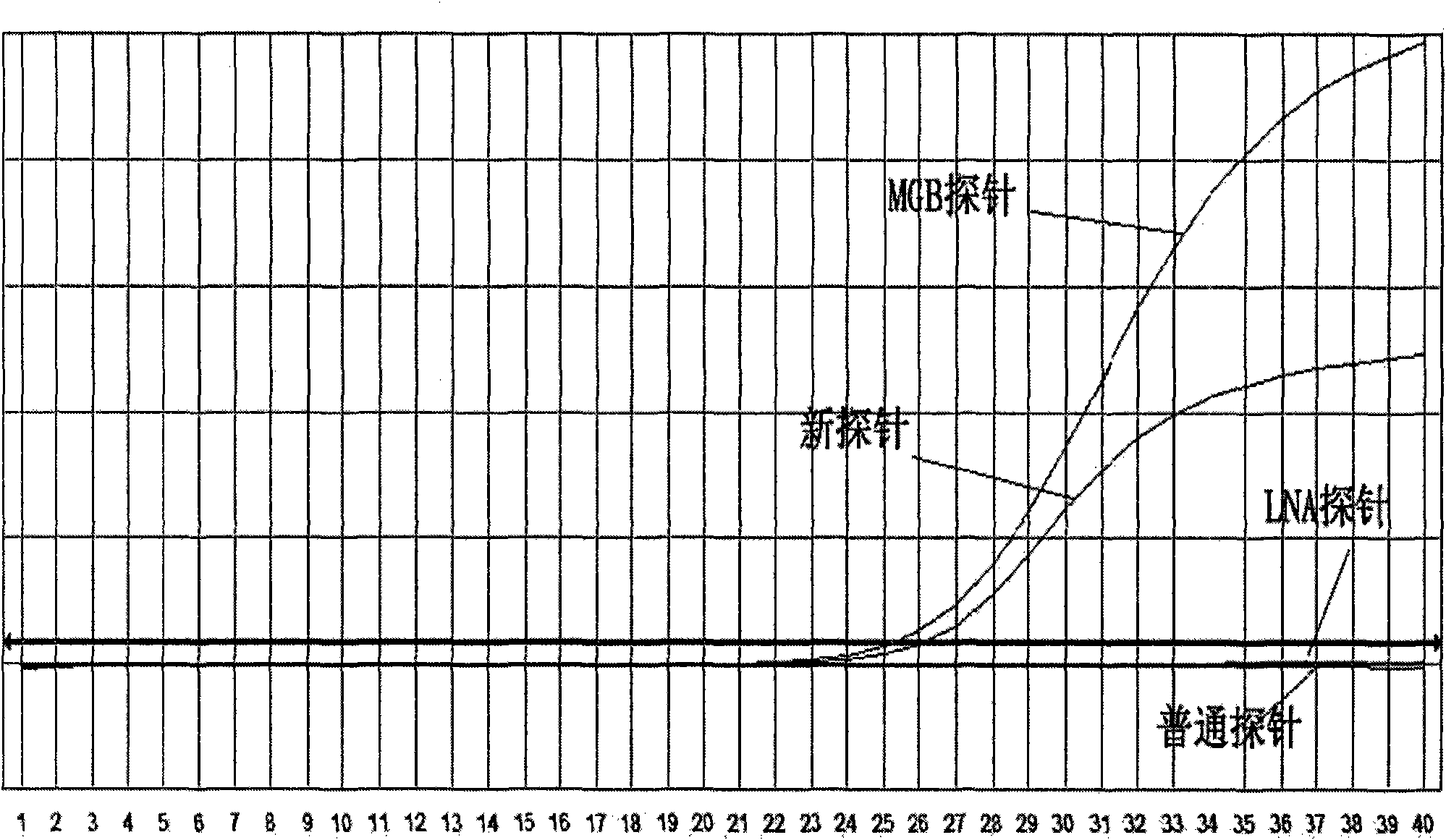
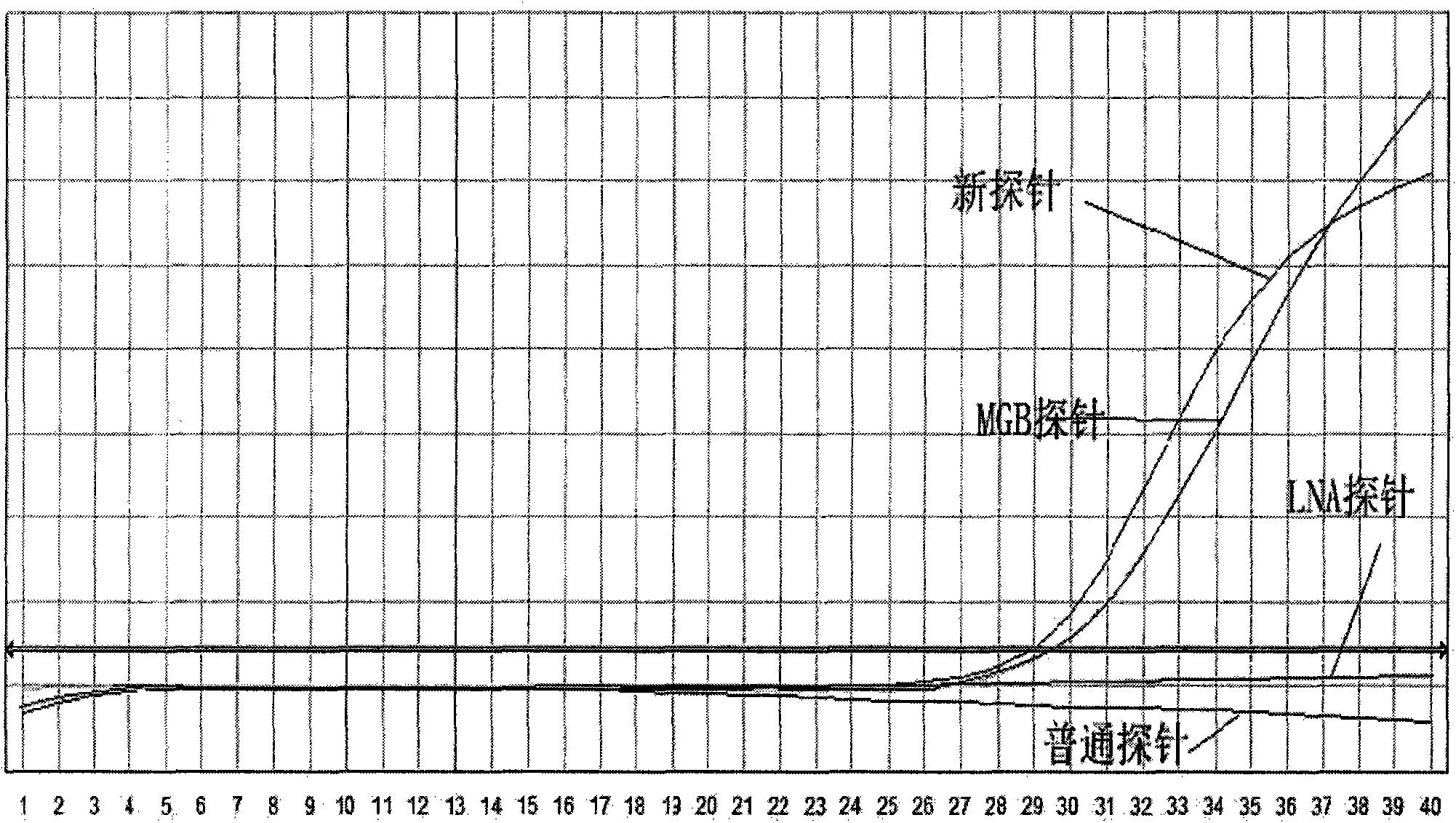
[0057] Embodiment 1: Take Taqman probe as an example
[0058] 1. Design and synthesis of primers and probes
[0059] Primers and probes were designed using the adefovir (ADV) mutation of hepatitis B virus as a detection template.
[0060] Design principles: 1) The primers are designed to amplify a nucleotide sequence containing adefovir (ADV) mutations according to the primer design principles.
[0061]2) The probe is designed with the mutant sequence as the target sequence.
[0062] The goal of the design: in hepatitis B positive samples, samples without adefovir (ADV) mutation cannot release fluorescent signals because the probe cannot bind to it; on the contrary, the mutant samples combine with the target nucleotide fragments to generate fluorescent signals .
[0063] Primer: Forward primer: ADV-236-1: 5'-TGGGCCTCAGTCCGTTTC-3' (SEQ ID NO: 1)
[0064] Reverse primer.: ADV-236-2: 5'-CCAATTACATATCCCATGAAGTTAAG-3' (SEQ ID NO: 2)
[0065] 1) common Taqman probe, 2) MGB prob...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com