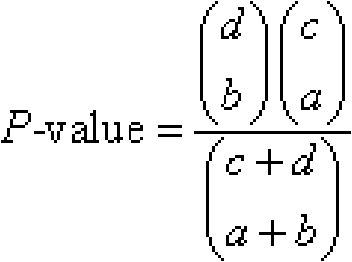Gene excavating method based on EST database and UniGene database
A database and gene technology, applied in the field of applied bioinformatics, can solve problems such as counting false positives of EST expression levels, inability to correctly mine induced genes, etc., and achieve the effects of avoiding errors, accurate analysis, and accurate results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
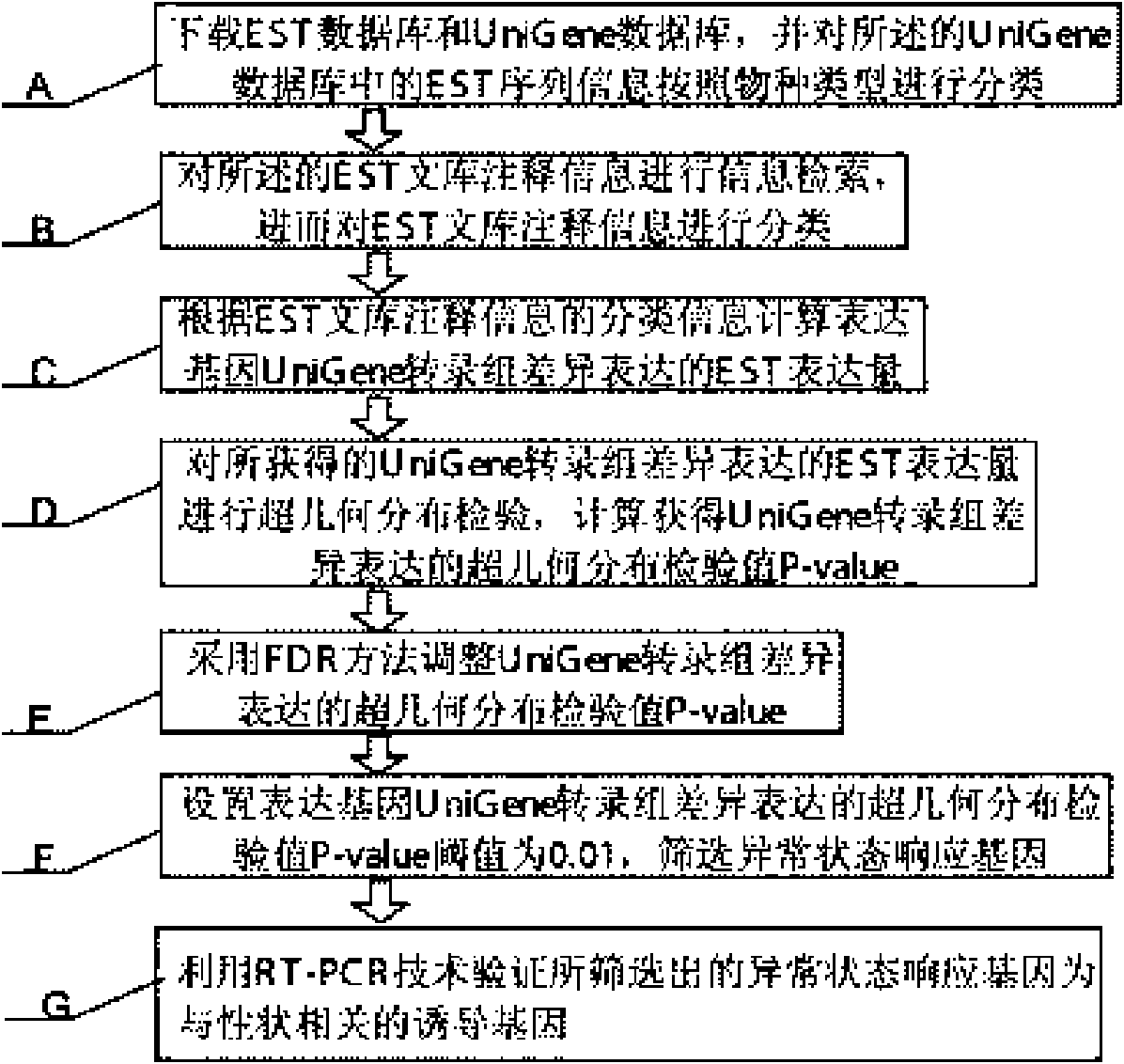
[0014] Specific embodiment one: see figure 1 The specific process of a gene mining method based on EST database and UniGene database described in this specific embodiment is:
[0015] Step A: Download the EST database and the UniGene database, and classify the EST sequence information in the UniGene database according to species type, and then perform step B;
[0016] Step B: Perform information retrieval on the annotation information of the EST library, and then classify the annotation information of the EST library, and then perform step C;
[0017] Step C: Calculate the EST expression level of the expressed gene UniGene transcriptome according to the classification information of the EST library annotation information, and then perform step D;
[0018] Step D: Perform hypergeometric distribution test on the EST expression of the obtained UniGene transcriptome of the expressed gene, calculate the hypergeometric distribution test value P-value of the differential expression of the exp...
specific Embodiment approach 2
[0033] Specific embodiment two: The gene mining method based on EST database and UniGene database described in this embodiment is different from specific embodiment one in that it is the mining of human cancer response genes:
[0034] In step A, download the human EST database and the human UniGene database;
[0035] In step B, the annotation information of the human EST library is directly extracted. First, the human abnormal state EST library is extracted, and the ID of the human abnormal state EST library is extracted, and then the human normal state EST library is extracted, and the human ID of the normal EST library;
[0036] In step C, according to the classification information of the annotation information of the human EST library, the health status of the human expression gene UniGene transcriptome is extracted from the human UniGene transcriptome file Hs.profiles in the human UniGene database. Breast tumor, leukemia under HealthState Leukemia, ovarian tumor, primitive neur...
specific Embodiment approach 3
[0244] Specific embodiment three: The gene mining method based on EST database and UniGene database described in this embodiment is different from specific embodiment one in that it is the mining of soybean stress response genes:
[0245] In step A, download the soybean EST database and soybean UniGene database;
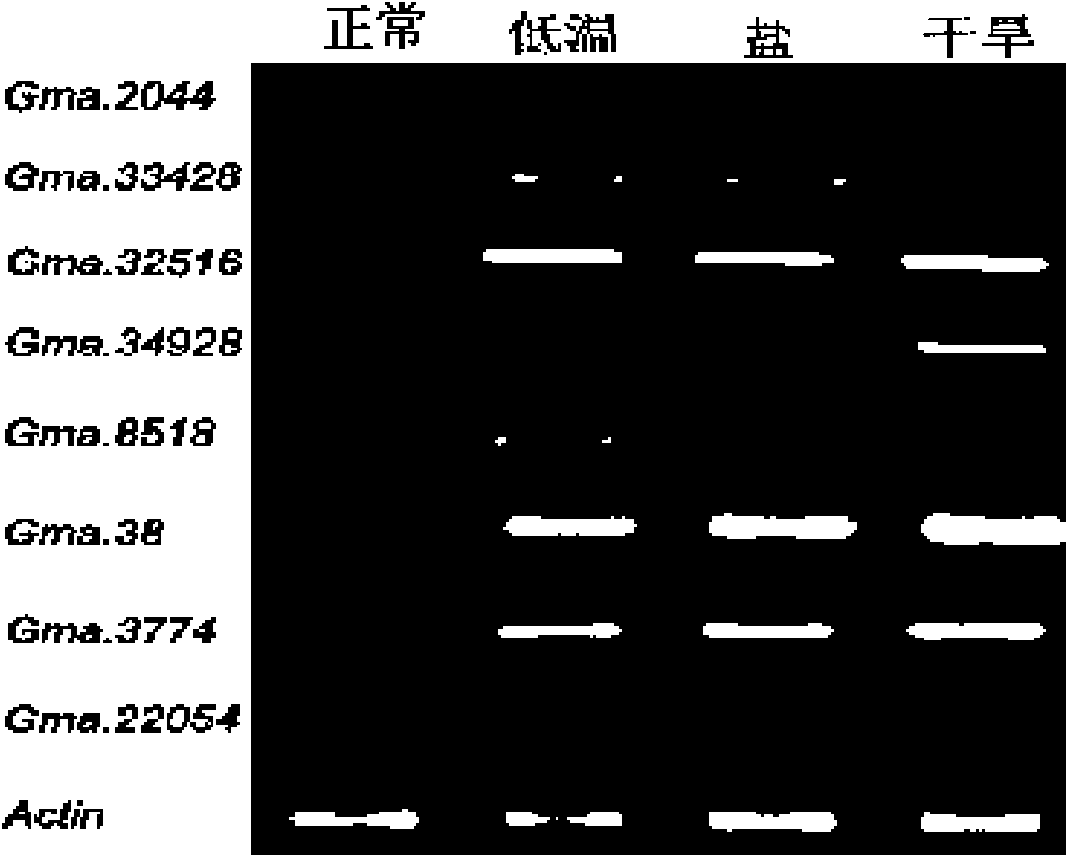
[0246] In step B, "cold", "salt" and "drought" are used as keywords of abnormal states related to soybean adversity stress, and the keywords are information in the soybean EST library annotation information file Gma.lib.info Search, screen the soybean abnormal state EST library, and extract the ID of the soybean abnormal state EST library, then use other EST libraries in the soybean EST library as the soybean normal state EST library, and extract the ID of the soybean normal state EST library. The search items for information retrieval in the soybean EST library annotation information file Gma.lib.info include three items: TITLE, DEVELOPMENTAL_STAGE and VERBATIM_TISSUE;
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com