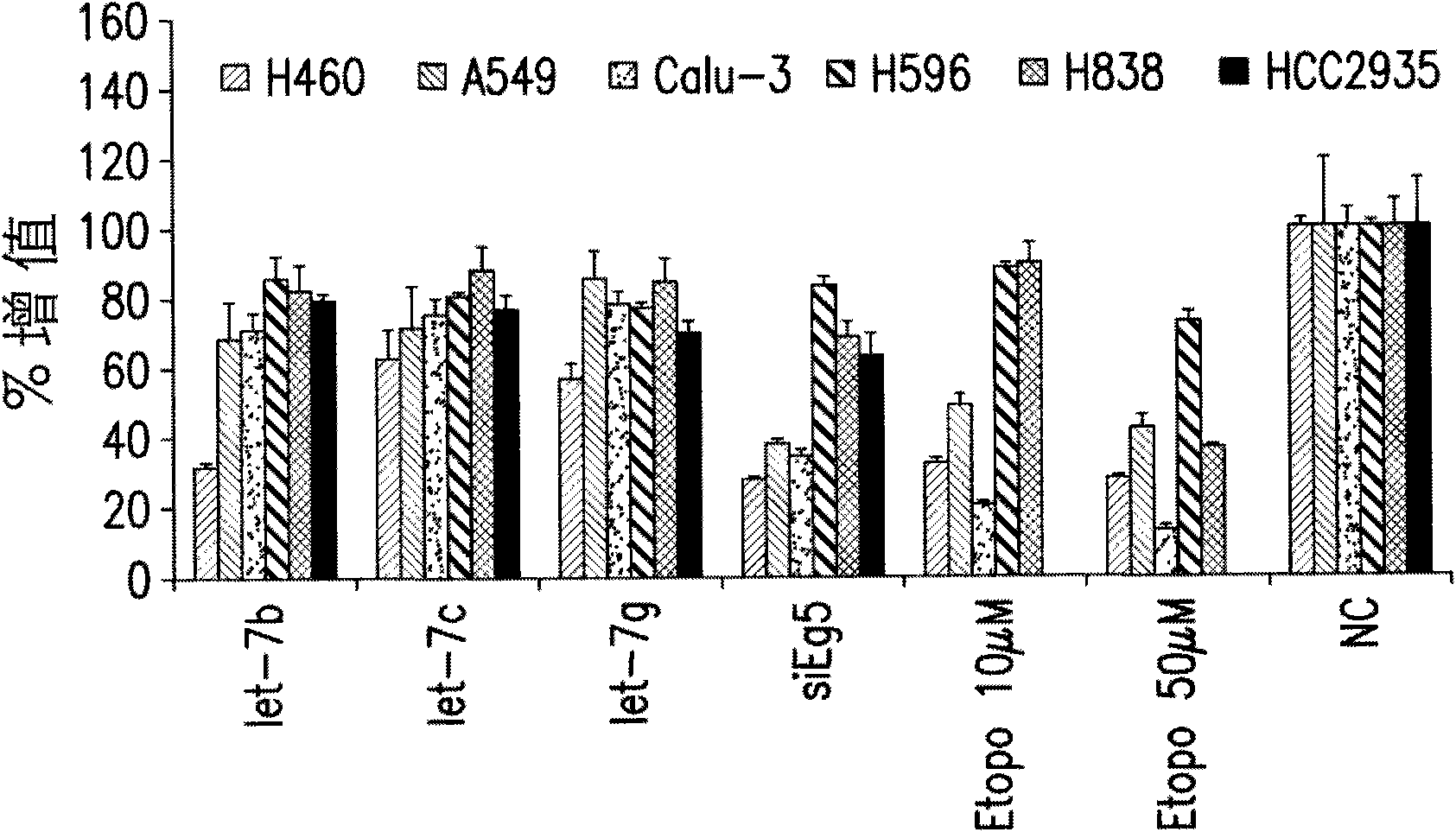
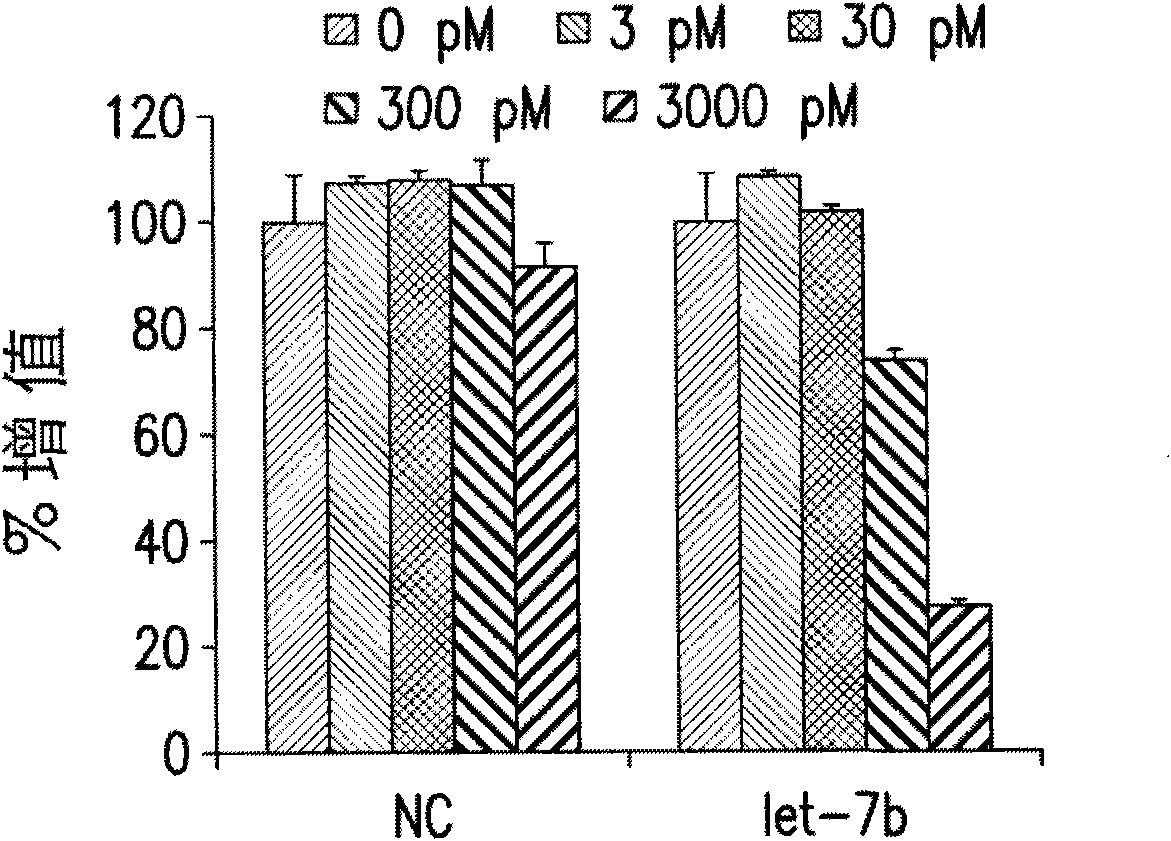
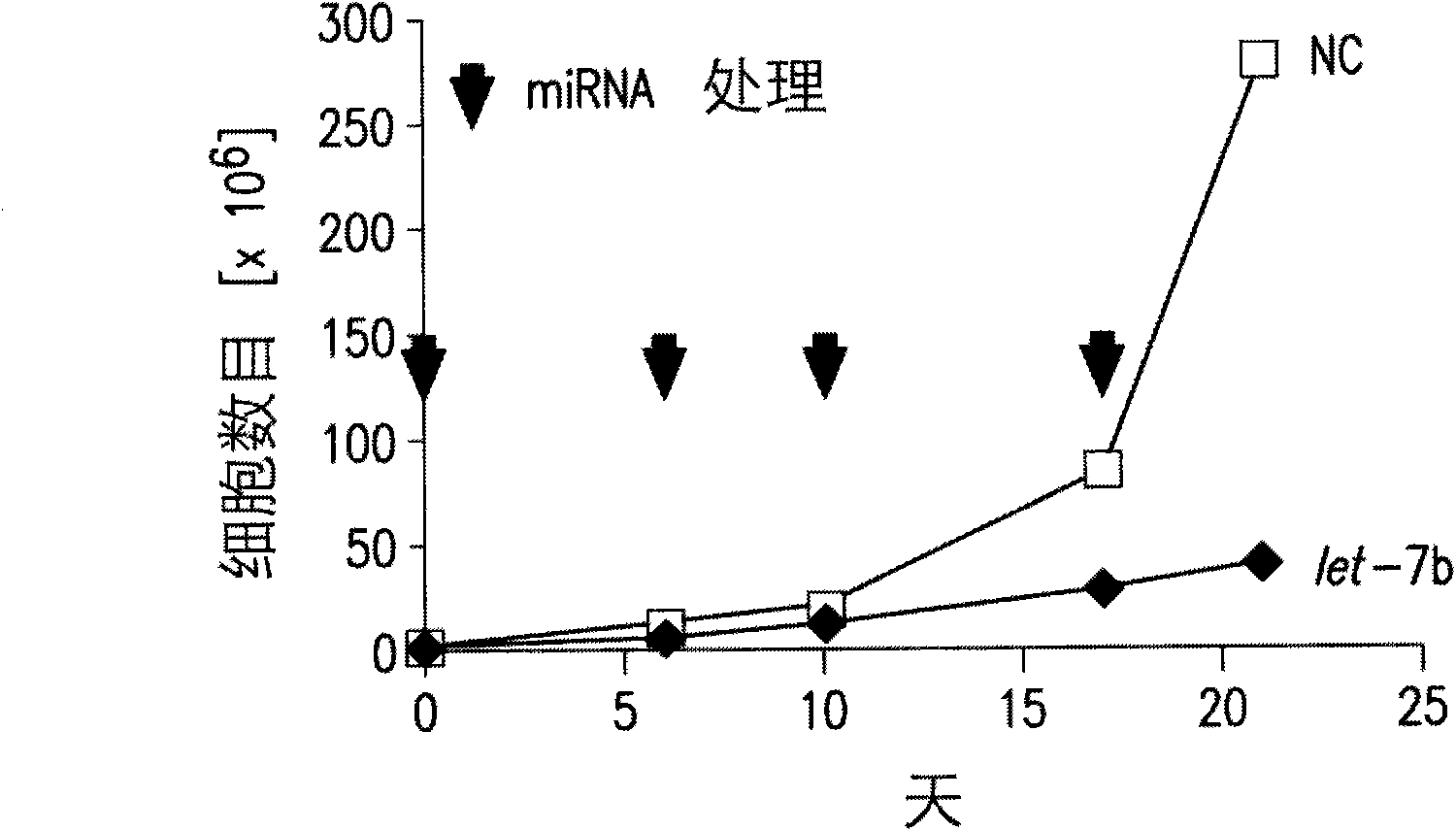
Functions and targets of let-7 micro rnas
A technology of let-7 and cells, applied in the field of molecular biology, can solve the problems of unknown details of regulatory pathways and networks, imprecise estimation of the number and identity of miRNA targets, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0241] B. Preparation of nucleic acid
[0242] Nucleic acids can be prepared by any technique known to those of ordinary skill in the art, such as chemical synthesis, enzymatic production or biological production. It is specifically considered that the miRNA probe of the present invention is chemically synthesized.
[0243] In some embodiments of the invention, miRNA is recovered or isolated from a biological sample. The miRNA can be recombinant, or it can be the cell's natural or endogenous miRNA (produced from the cell's genome). It is considered that biological samples can be processed to enhance the recovery of small RNA molecules such as miRNA. This method is described in US Patent Application Serial No. 10 / 667,126, which is specifically incorporated herein by reference. Generally, these methods involve lysing cells with a solution with guanidine salt and detergent.
[0244] Alternatively, nucleic acid synthesis can be performed according to standard methods. See, for examp...
Embodiment 1
[0285] Method for analyzing gene expression after miRNA transfection
[0286] The synthetic precursor miR miRNA (Ambion) was reverse-stained into quadruplicate samples of A549, HepG2 or HL-60 cells. A549 and HepG2 cells were transfected with siPORT NeoFX (Ambion) according to the manufacturer's recommendation using the following parameters: 200,000 cells per well in a 6-well plate, 5.0 μl NeoFX, 2.5 ml of miRNA at a final concentration of 30 nM. The cells were harvested 72 hours after transfection. HL-60 cells were transfected with the Gene Pulser Xcell System (Bio-Rad Laboratories, Inc.; Hercules, CA, USA) according to the manufacturer’s instructions and the following parameters: 500,000 cells in a volume of 150 μl per reaction, in a 2-mm electroporation cuvette (electroporation cuvette), a single square wave pulse of 25msec is used.
[0287] Total RNA was extracted with RNAqueous-4PCR (Ambion) according to the manufacturer's recommended protocol. The mRNA array analysis was p...
Embodiment 2
[0289] Gene expression analysis of A549, HEPG2 and HL-60 cells transfected with HSA-LET-7B
[0290] miRNAs are thought to affect gene expression mainly at the translation level. However, it has recently been reported that in some cases hsa-let-7 (Bagga et al., 2005) and other miRNAs (Lim et al., 2005) can reduce the mRNA level of direct targets, and this change can be used in microarray gene expression. Observed during analysis.
[0291] The experiments described herein can identify genes whose mRNA levels are affected by hsa-let-7 expression in human lung cancer (A549) cell lines, human liver cancer cell lines (HepG2), and human acute myeloid leukemia cell lines (HL-60). A549, HepG2 or HL-60 cells were transfected with the precursor miR hsa-let-7b (as a representative member of the hsa-let-7 miRNA family) as described in Example 1. The results of microarray gene expression analysis are shown in Tables 2, 3, and 4.
[0292] Table 2. Genes with altered mRNA expression levels in A5...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com