Molecular mark SIsvo188 closely linked with herbicide-resistant gene in foxtail millet
A herbicide-resistant gene and molecular marker technology, applied in the field of molecular biology, can solve the problems of lack of herbicide-resistant, large quantity, and obstacles to millet production, and achieve the effect of high-throughput application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: Construction of millet F2 generation segregation population
[0044] The male parent is Zhanggu No. 1: Anti-Nabujing type (high plant type, plant height is about 150cm), long and narrow flag leaves, red bristles, red glumes, fertile, greenish leaf color.
[0045] The female parent is the A2 sterile line: non-resistant Nabujing type (short plant type, plant height about 100cm), short and wide flag leaves, green bristles, green glumes, partly sterile, yellowish leaf color.
[0046] F1 (Zhang Zagu No. 3, partially anti-Nabu net type, plant height of about 130cm) was obtained by crossing the male parent and the female parent.
[0047] The F1 generation was self-crossed to generate the F2 generation population, and a total of 480 individual plants were obtained. 480 individuals of the F2 generation were analyzed according to the method for determining the type of anti-Natura, and a total of 360 strains were found to be anti-Natten and partly anti-Nart (includin...
Embodiment 2
[0048] Example 2: Extraction of parental and F1 generation, F2 generation individual genomic DNA
[0049] Genomic DNA of the parents, the F1 generation, and 480 F2 generation individuals in Example 1 were extracted by the CTAB method, and the specific methods were as follows:
[0050] (1) Weigh 1.0g of fresh leaves, cut them into pieces and put them in a mortar, grind them with liquid nitrogen, add 3ml of 1.5×CTAB, grind them into a homogenate and transfer them to a 15ml centrifuge tube, then add 1ml of 1.5× CTAB into the mortar Rinse with CTAB and transfer to a centrifuge tube. After mixing, place in a water bath at 65°C for 30 minutes, and shake slowly from time to time.
[0051] Among them, the formula of 1.5×CTAB is as follows (1L):
[0052] CTAB 15g
[0053] 1M Tris.Cl (pH 8.0) 75ml
[0054] 0.5M EDTA 30ml
[0055] NaCl 61.4g
[0056] Add deionized water to make up to 1 L, and add mercaptoethanol with a final concentration of 0.2% (2 ml) before use.
[0057] (2) ...
Embodiment 3
[0062] Example 3: Preparation of Molecular Markers
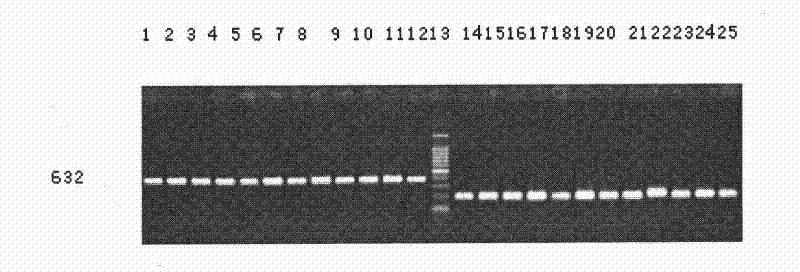
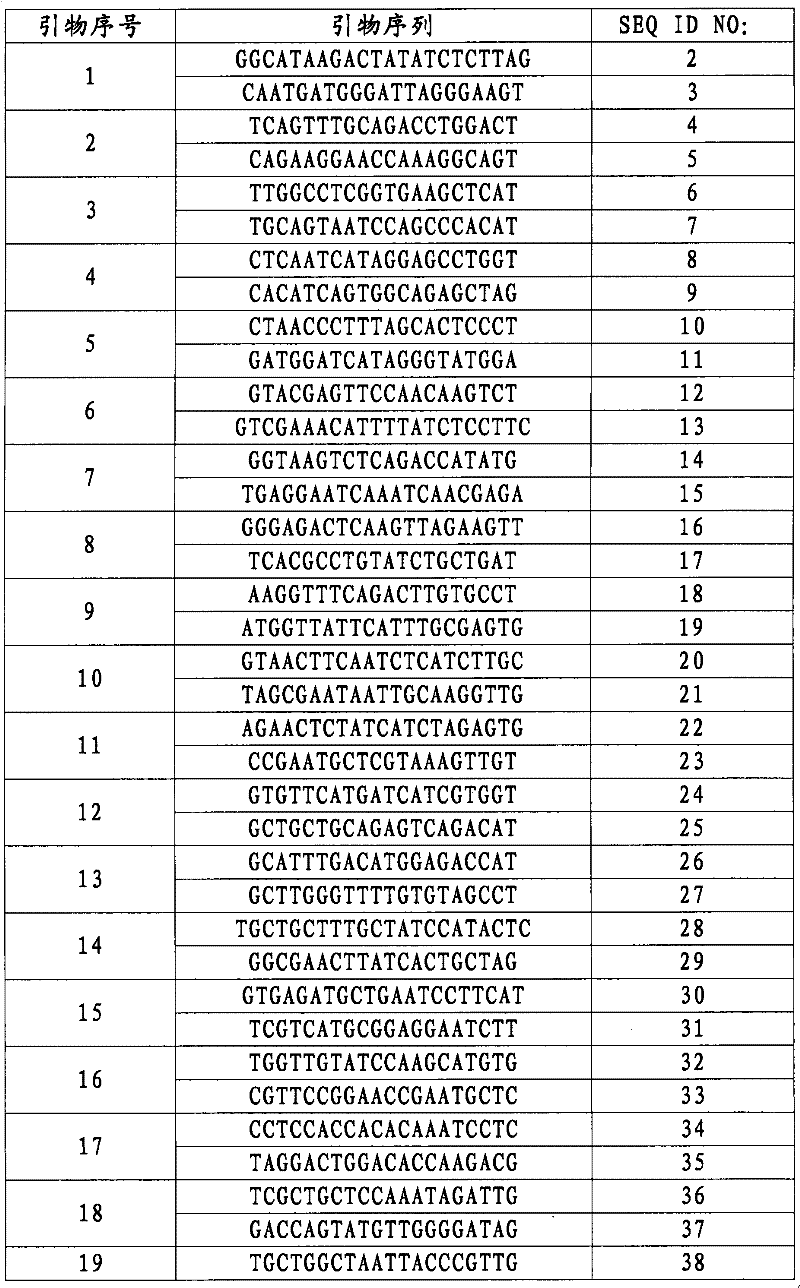
[0063] Taking the Genomic DNA of the anti-Narth net type or part of the anti-Natur net type in the male parent, F1 generation, or F2 generation extracted in Example 2 as a template, with molecular marker primers (SEQ ID NO: 2 and SEQ ID NO: 3 ) for PCR amplification.
[0064] The PCR reaction system is as follows:
[0065] Sterile water 20.2μl
[0066] 10*Buffer (with Mg 2+ ) 2.5 μl
[0067] dNTPs (25mM) 0.15μl
[0068] Taq enzyme (5U / μl) 0.15μl
[0069] Forward primer 0.5μl
[0070] Reverse primer 0.5μl
[0071] Template 1.0μl
[0072] Total volume 25μl
[0073] The PCR reaction procedure is as follows:
[0074] Pre-denaturation at 94°C for 5 minutes; denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, extension at 72°C for 40 seconds, and 35 cycles; final extension at 72°C for 3 minutes. PCR amplification products can be stored at 4°C.
[0075] The amplified product is purified to obtain mo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com