Multiple detection method for DNA polymorphism of genome and special probe thereof
A polymorphism and genome technology, applied in the field of genetic engineering, can solve the problem of low throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0110] Embodiment 1, the button hand probe of single nucleotide polymorphism (SNP)
[0111] 1. Detection of two SNPs in the genome of Aegilops tauschii
[0112] 1. Design of button probes PdPCJ819266A, PdPCJ819266G, PdPBJ262615T, PdPBJ262615C
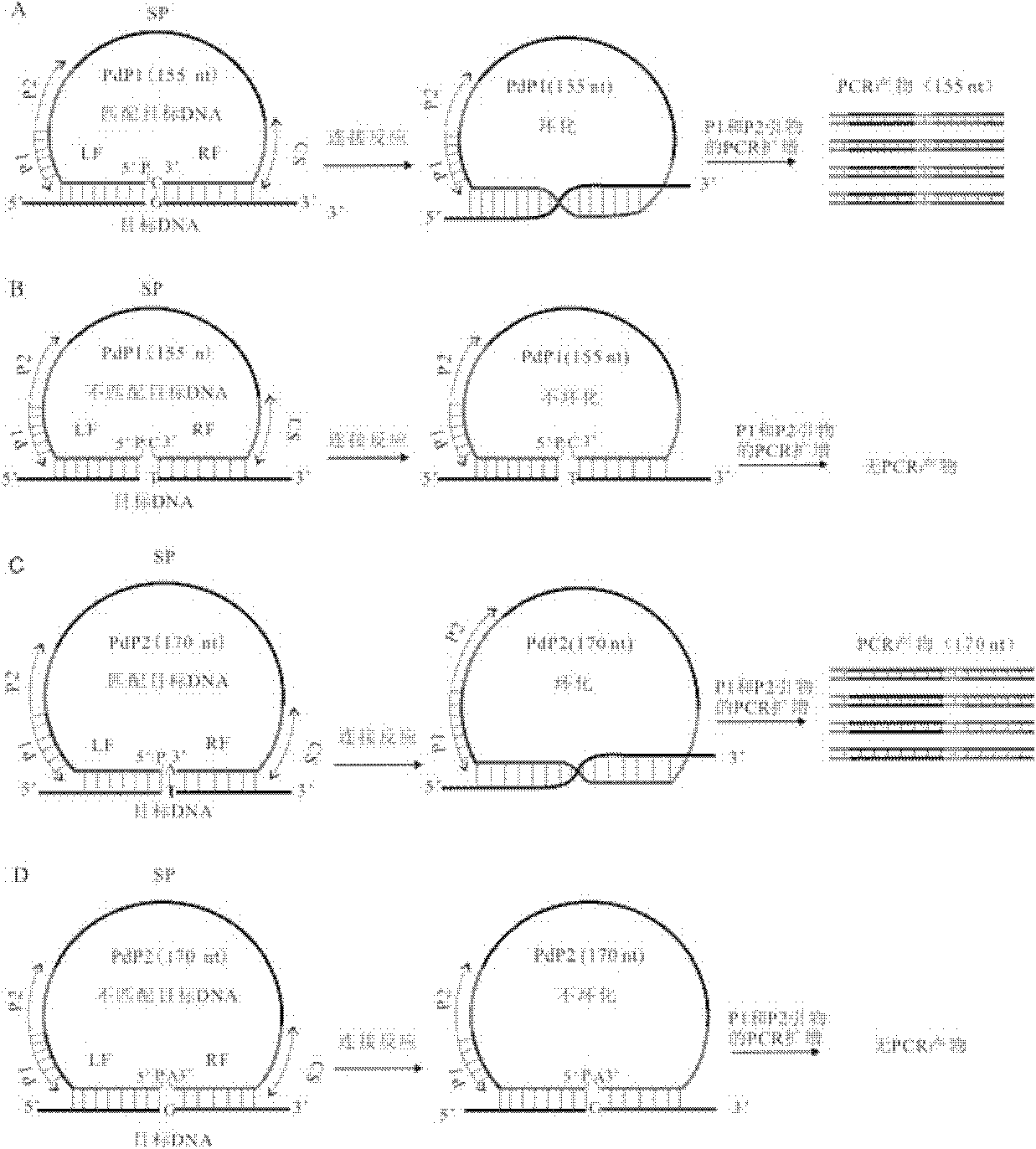
[0113] The genome of Aegilops tauschii is the donor of the D genome of common wheat (Triticum aestivum). PdPCJ819266 and PdPBJ262615 are two SNPs of Aegilops tauschii (Aegilops tauschii), in which AA and TT are genotypes of Aegilops tauschii variety 2280, and GG and CC are genotypes of Aegilops tauschii variety 2282, that is, Aegilops tauschii 2280 and Aegilops tauschii The 62nd nucleotide in the sequence 5 of the 2282 variety has a polymorphism, which are AA and GG respectively; the 175th nucleotide in the sequence 6 of the 2280 and 2282 varieties of A. tachyphylla has a polymorphism, which are respectively TT and CC.
[0114] Among them, the button-hand probe PdPCJ819266A specifically recognizes allele A with a length of 147 nt, th...
Embodiment 2
[0161] Embodiment 2, insertion / deletion polymorphism (InDel) labeled button hand probe
[0162] 1. Detection of two insertion / deletion (InDel) markers in the barley genome
[0163] 1. Design of button probes PdPInDel1-1, PdPInDel1-2, PdPInDel2-1, PdPInDel2-2
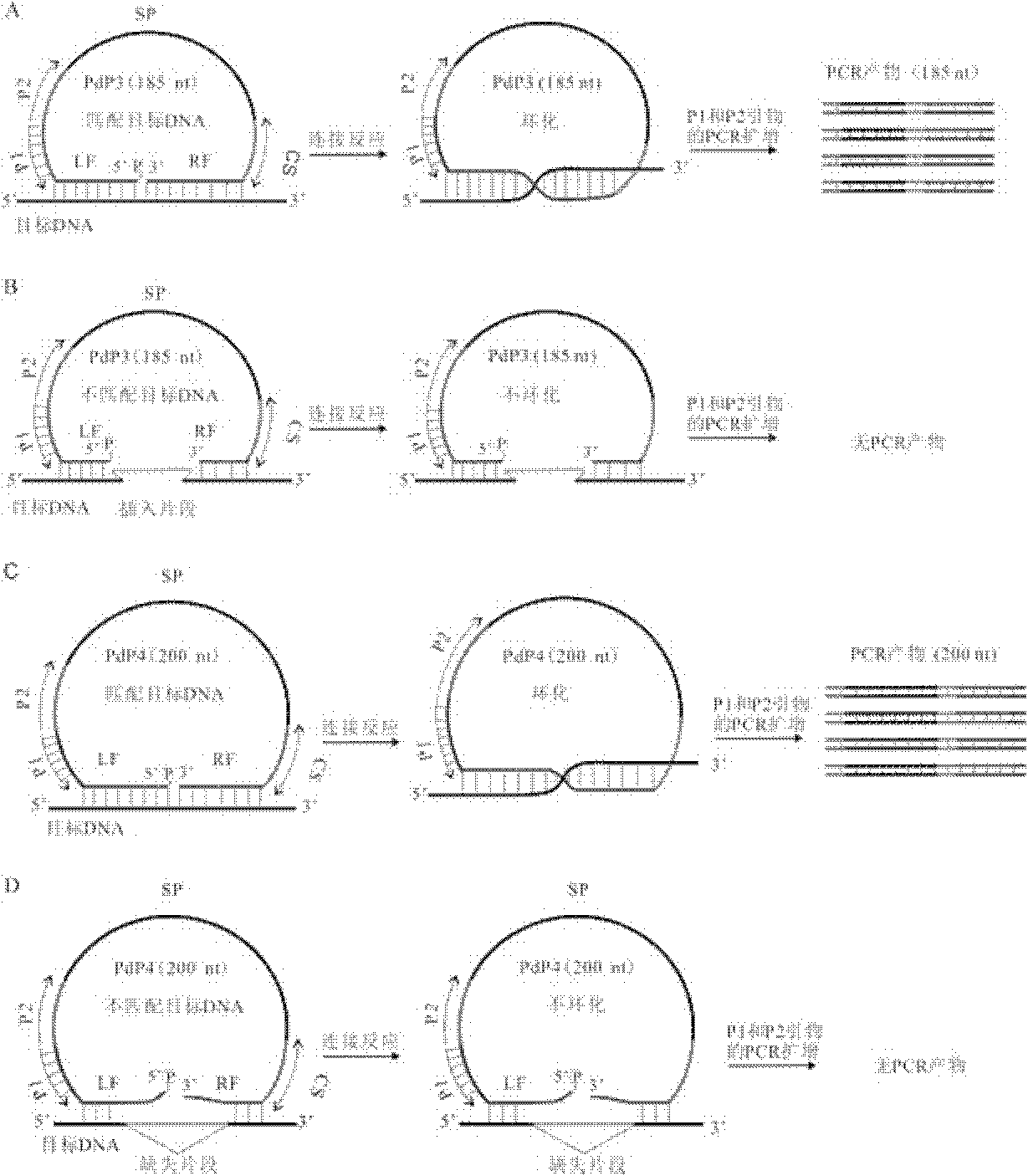
[0164] PdPInDel1 and PdPInDel2 are two InDel markers on the fifth chromosome (5H) of barley (Hordeum vulgare L.), wherein InDel1-1 and InDel2-1 are specific genotypes of barley variety Vada, and InDel1-2 and InDel2-2 are Genotypes specific to barley cultivar L94. The button-hand probe PdPInDel1-1 specifically recognizes the InDel1-1 genotype, and its length is 160 nt. The button-hand probe PdPInDel1-2 specifically recognizes the InDel1-2 genotype, and its length is 190 nt. The genotype is 318nt in length, and the clasp probe PdPInDel2-2 specifically recognizes the InDel2-2 genotype with a length of 342nt.
[0165] The 5' ends of the four hand probes were all phosphorylated. See Table 4 for sequence design and structur...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com