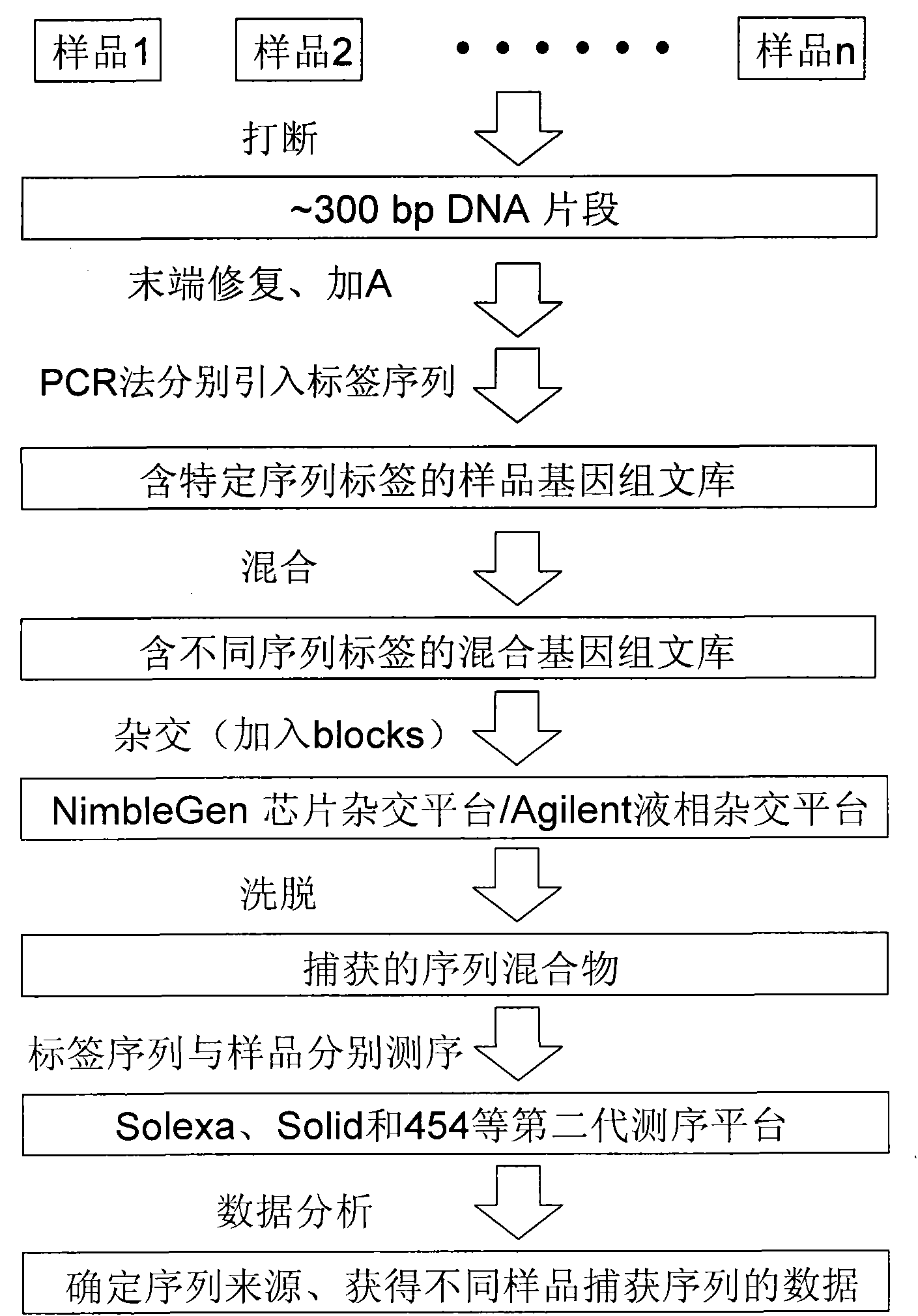
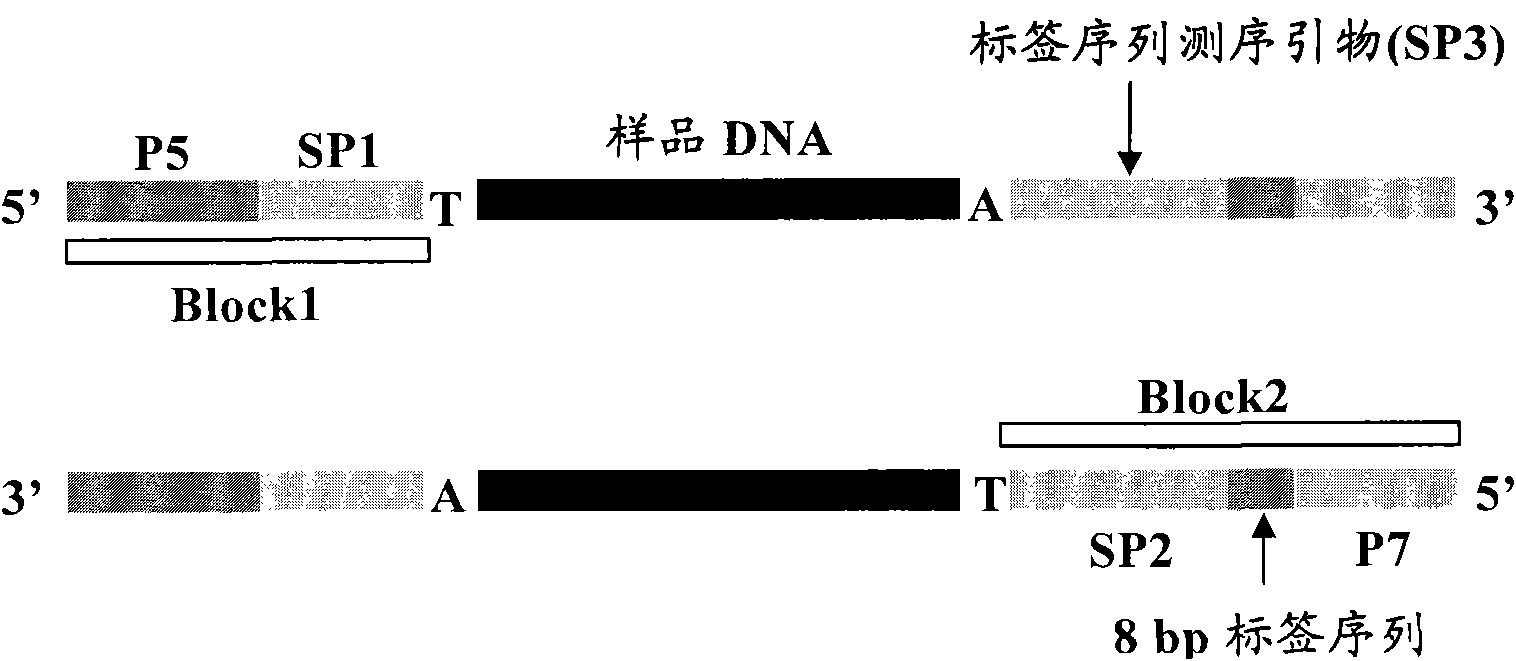
Method for building sequencing library by hybridization
A library and genome library technology, applied in hybridization-based sequence capture technology, to achieve sequence capture of multiple samples in the same reaction system, second-generation sequencing technology, solexa sequencing technology, labeling technology, can solve the waste of sequencing data , poor sequence capture effect, non-target sequences, etc., to avoid data waste, avoid the reduction of sequencing length, and improve the efficiency of introduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] The control example of embodiment 1.NimbleGen chip hybridization system (Roche NimbleGen company): Single sample is hybridized on Nimblegen 855K chip
[0085] (1) Experimental method:
[0086] 1. Hybrid library construction
[0087] Refer to the Illumina Multiplexing Sample Preparation Guide for the construction process of the hybridization library. After 3ug of genomic DNA (extracted from human peripheral blood) was interrupted, the ends were blunted, "A" bases were added, and adapters (from the Illumina Multiplexing Sample Preparation Oligonucleotide Kit) were added and carried out. PCR amplification, PCR reaction system and reaction conditions are as follows:
[0088] reaction system:
[0089]
[0090] Reaction conditions:
[0091] (a).98℃ 30s
[0092] (b).98℃ 15s
[0093] (c).65℃ 30s
[0094] (d).72℃ 30s
[0095] (e). Repeat steps (b)-(d) 3-9 times (4-10 cycles in total)
[0096] (f).72℃ for 5 minutes
[0097] (g). Stand at 4℃
[0098] Use Ampure beads ...
Embodiment 2
[0126] Example 2. Example of application in the NimbleGen chip hybridization system: 12 DNA libraries (constructed according to the hybridization library construction process) are mixed and then sequence captured with an 855K chip
[0127] (1) Experimental method:
[0128] 1. Preparation of hybrid library: the method is the same as that described in Example 1.
[0129] 2. Hybridization:
[0130] a. Sample preparation:
[0131]
[0132] The 12 libraries (constructed according to the hybridization library construction method) were mixed together and hybridized on the same chip, and the hybridization method was the same as in Example 1.
[0133] 3. Sequencing and data analysis: the method is the same as that described in Example 1.
[0134] (2) Results:
[0135] After the 12 samples were mixed, the effect of sequence capture with the 855K chip is shown in Table 2.
[0136] Table 2: Sequence capture effect with 855K chip after mixing 12 samples:
[0137]
Embodiment 3
[0138] Example 3. Example of application in the NimbleGen chip hybridization system: 24 libraries (constructed according to the hybridization library construction method) are mixed and then sequence captured with an 855K chip
[0139] (1) Experimental method:
[0140] 1. Preparation of hybrid library: the method is the same as in Example 1.
[0141] 2. Hybridization:
[0142] a. Sample preparation:
[0143]
[0144] 24 samples were mixed together and hybridized on the same chip, and the hybridization method was the same as in Example 1.
[0145] 3. Sequencing and data analysis: the method is the same as in Example 1.
[0146] (2) Results:
[0147] The effect of sequence capture with 855K chip after mixing 24 samples is shown in Table 3.
[0148] Table 3: Sequence capture effect with 855K chip after mixing 24 samples:
[0149]
[0150] Summary of the implementation results of the NimbleGen chip hybridization system: In Examples 2 and 3, 12 and 24 samples were respec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com