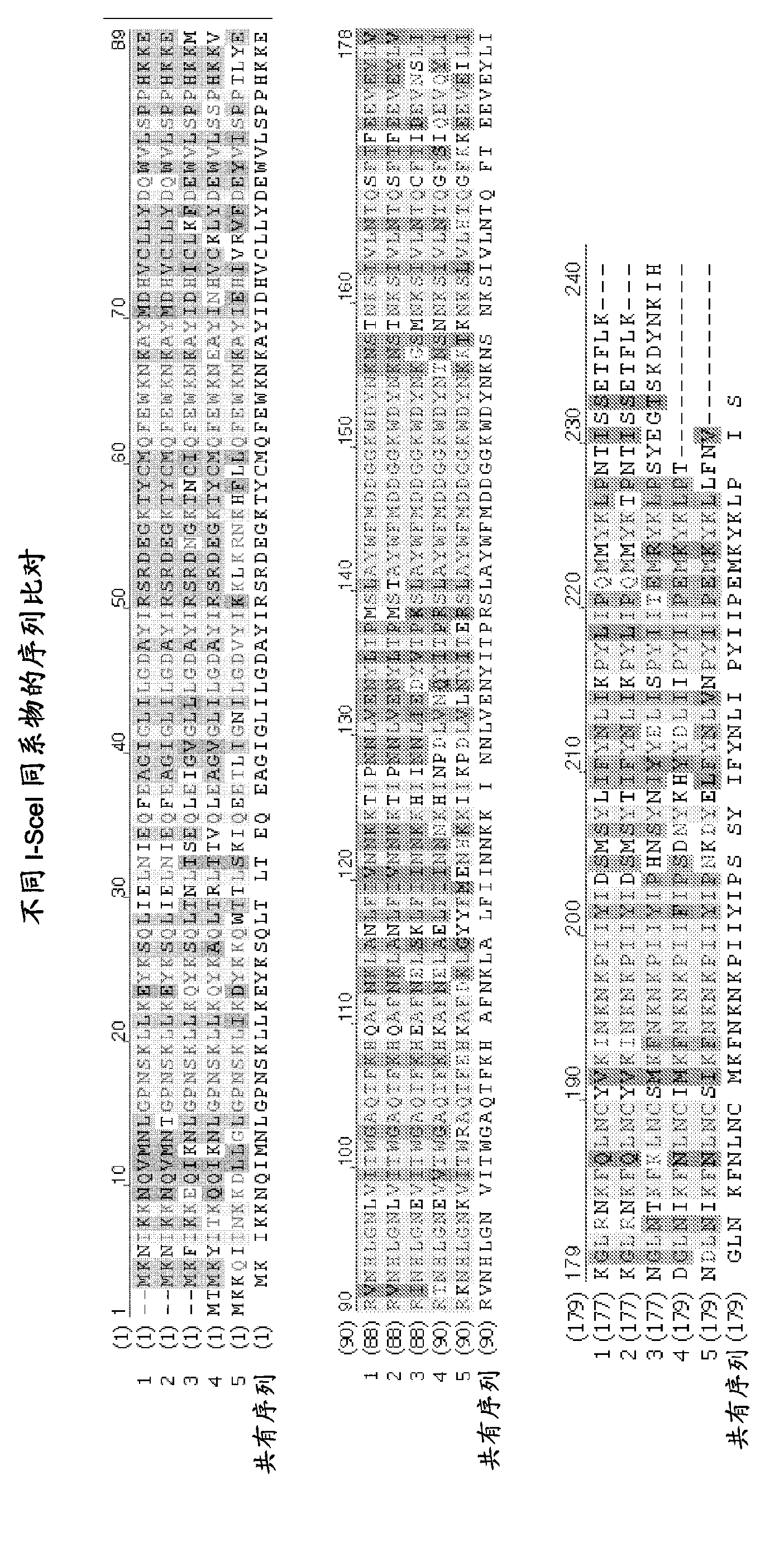
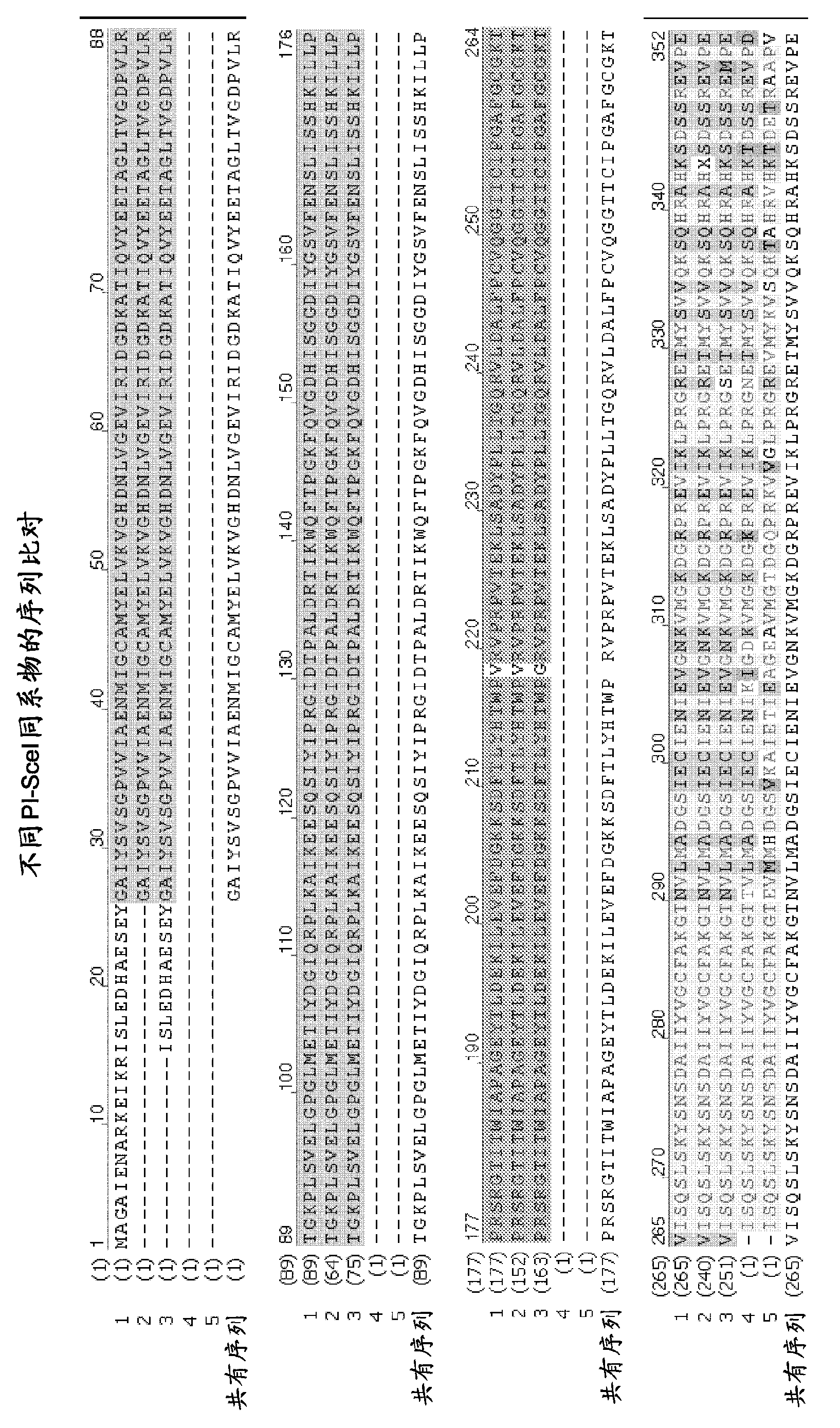
Chimeric endonucleases and uses thereof
A technology of endonuclease and polynucleotide, applied in the field of chimeric endonuclease, can solve problems such as limiting off-target effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0365] Preparation of Chimeric Endonucleases
[0366] Endonucleases and heterologous DNA binding domains can be combined in a number of alternative ways.
[0367] For example, more than one endonuclease can be combined with one or more heterologous DNA binding domains, or more than one heterologous DNA binding domain can be combined with one endonuclease. It is also possible to combine more than one endonuclease with more than one heterologous DNA binding domain.
[0368] The heterologous DNA binding domain can be fused to the N-terminal end or the C-terminal end of the endonuclease. One or more heterologous DNA binding domains can also be fused to the N-terminal end of the endonuclease and one or more heterologous DNA binding domains can be fused to the C-terminal end of the endonuclease. Alternating combinations of endonucleases and heterologous DNA binding domains can also be made.
[0369] Where chimeric endonucleases contain more than one endonuclease or more than on...
Embodiment 1
[0604] Example 1: Constructs with sequence-specific DNA endonuclease expression cassettes for expression in E. coli
Embodiment 1a
[0605] Example 1a: Basic constructs
[0606] In this example, we show a general outline of a vector designated "Construct I", which is suitable for transformation in E. coli. This general outline of the vector contains the ampicillin resistance gene for selection, the origin of replication for E. coli and the gene araC (which encodes an arabinose-inducible transcriptional regulator). Different genes encoding different versions of sequence-specific DNA endonucleases can be expressed from the arabinose-inducible pBAD promoter (Guzman et al., J Bacteriol 177:4121-4130 (1995)). Gene sequences encoding different nuclease versions are given in the Examples below.
[0607] The control construct in which the sequence encoding I-SceI (SEQ ID NO: 22) was designated VC-SAH40-4.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com